Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
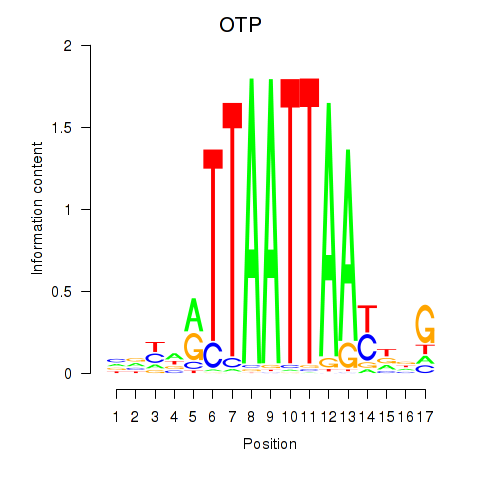
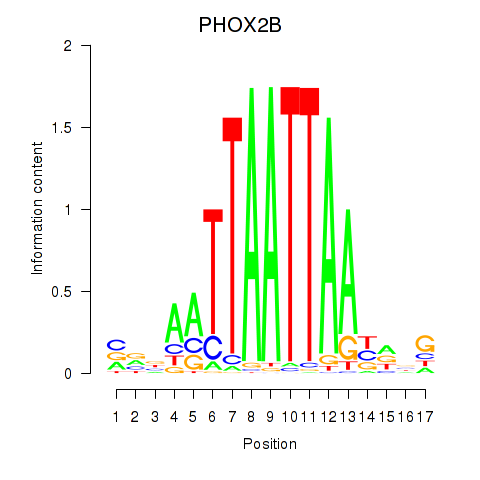
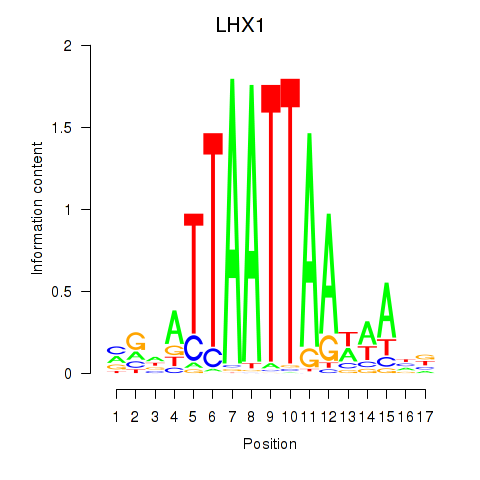
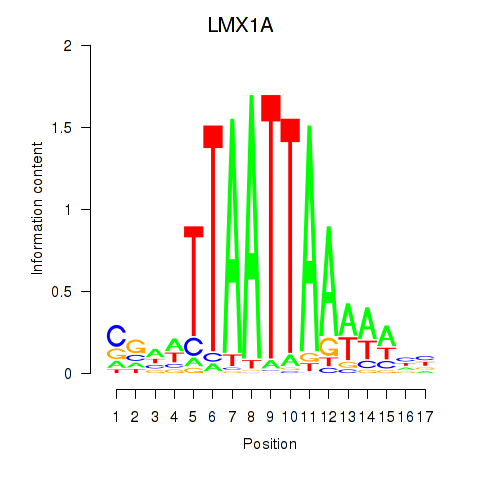
Results for OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Z-value: 0.84
Transcription factors associated with OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTP
|
ENSG00000171540.6 | orthopedia homeobox |
|
PHOX2B
|
ENSG00000109132.5 | paired like homeobox 2B |
|
LHX1
|
ENSG00000132130.7 | LIM homeobox 1 |
|
LMX1A
|
ENSG00000162761.10 | LIM homeobox transcription factor 1 alpha |
|
LHX5
|
ENSG00000089116.3 | LIM homeobox 5 |
|
HOXC4
|
ENSG00000198353.6 | homeobox C4 |
|
HOXC4
|
ENSG00000273266.1 | homeobox C4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC4 | hg19_v2_chr12_+_54447637_54447661 | 0.67 | 1.5e-01 | Click! |
| LHX1 | hg19_v2_chr17_+_35294075_35294102 | -0.15 | 7.7e-01 | Click! |
| OTP | hg19_v2_chr5_-_76935513_76935513 | 0.11 | 8.3e-01 | Click! |
Activity profile of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
Sorted Z-values of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_327537 | 1.49 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr14_+_20187174 | 1.27 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr10_+_91152303 | 0.79 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr17_+_1674982 | 0.71 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr5_-_126409159 | 0.68 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr11_-_121986923 | 0.48 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr1_+_66820058 | 0.46 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr10_+_94451574 | 0.43 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr19_-_36304201 | 0.42 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr2_+_182850551 | 0.41 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_182850743 | 0.40 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr3_-_191000172 | 0.39 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr1_+_152178320 | 0.39 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr16_-_1538765 | 0.38 |
ENST00000447419.2
ENST00000440447.2 |
PTX4
|
pentraxin 4, long |
| chr3_+_138340049 | 0.38 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_+_113568207 | 0.37 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr12_+_12510352 | 0.36 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr16_-_30122717 | 0.36 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr17_-_38821373 | 0.34 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr6_-_41039567 | 0.33 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr4_+_155484103 | 0.33 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484155 | 0.31 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr7_+_5465382 | 0.30 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr8_-_90769422 | 0.29 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chrX_+_43515467 | 0.29 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr3_-_122283424 | 0.28 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr6_+_130339710 | 0.28 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr2_-_37068530 | 0.27 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr4_-_39033963 | 0.26 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr12_-_112123524 | 0.26 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr12_+_104337515 | 0.25 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chrX_+_51486481 | 0.25 |
ENST00000340438.4
|
GSPT2
|
G1 to S phase transition 2 |
| chr2_+_152214098 | 0.24 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr12_+_78359999 | 0.24 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr7_+_133812052 | 0.23 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr6_+_26440700 | 0.23 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr7_-_101272576 | 0.22 |
ENST00000223167.4
|
MYL10
|
myosin, light chain 10, regulatory |
| chr17_+_19091325 | 0.22 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr8_-_124741451 | 0.22 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr17_+_48823975 | 0.21 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr19_+_50016411 | 0.21 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr1_+_62439037 | 0.21 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr10_-_99030395 | 0.20 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr3_+_23851928 | 0.20 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr2_-_43266680 | 0.20 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr12_-_54653313 | 0.20 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chrX_+_84258832 | 0.20 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr18_-_32870148 | 0.20 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr4_+_95128748 | 0.19 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr14_-_54425475 | 0.18 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr12_+_64798095 | 0.18 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr4_+_76481258 | 0.17 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr6_+_155334780 | 0.17 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr18_+_47087390 | 0.17 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr5_+_179135246 | 0.17 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr7_+_138915102 | 0.17 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr7_-_87342564 | 0.17 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chrX_-_21676442 | 0.17 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr11_-_107729504 | 0.16 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_-_177116772 | 0.16 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr11_+_5710919 | 0.16 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr2_+_191221240 | 0.16 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr4_+_119809984 | 0.16 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr9_-_99540328 | 0.16 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr2_-_40680578 | 0.16 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr16_-_66583994 | 0.15 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr3_+_139063372 | 0.15 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr12_-_118796971 | 0.15 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr1_+_117544366 | 0.15 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr11_+_35222629 | 0.15 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr4_+_86525299 | 0.15 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_+_57233087 | 0.15 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr11_+_71934962 | 0.15 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr3_-_141747950 | 0.14 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_-_74618907 | 0.14 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chrM_+_14741 | 0.14 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chrX_+_10126488 | 0.14 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr16_-_28634874 | 0.14 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr3_+_44840679 | 0.13 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr3_-_141719195 | 0.13 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_212965104 | 0.13 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr17_+_68164752 | 0.13 |
ENST00000535240.1
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr19_-_36001113 | 0.13 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr5_+_89770664 | 0.13 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr12_+_4130143 | 0.13 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chr9_-_26947220 | 0.12 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
| chr8_+_1919481 | 0.12 |
ENST00000518539.2
|
KBTBD11-OT1
|
KBTBD11 overlapping transcript 1 (non-protein coding) |
| chr5_+_66300446 | 0.12 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_212965170 | 0.12 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr17_-_19015945 | 0.11 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr11_-_107729287 | 0.11 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_+_158493642 | 0.11 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr18_-_47018869 | 0.11 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr19_-_44388116 | 0.11 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr12_+_26164645 | 0.11 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr21_-_15583165 | 0.10 |
ENST00000536861.1
|
LIPI
|
lipase, member I |
| chr4_+_26324474 | 0.10 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr15_+_69857515 | 0.10 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr4_+_119810134 | 0.10 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr12_-_10282742 | 0.10 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr3_+_138340067 | 0.09 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr5_+_89770696 | 0.09 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr3_+_45927994 | 0.09 |
ENST00000357632.2
ENST00000395963.2 |
CCR9
|
chemokine (C-C motif) receptor 9 |
| chr18_-_47018897 | 0.09 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr16_-_66583701 | 0.09 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr3_-_33686925 | 0.09 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr7_-_64023441 | 0.09 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr17_-_42992856 | 0.09 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr9_-_26947453 | 0.09 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr10_-_28571015 | 0.09 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_+_75784850 | 0.08 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr1_+_151253991 | 0.08 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr18_+_47087055 | 0.08 |
ENST00000577628.1
|
LIPG
|
lipase, endothelial |
| chr10_-_25304889 | 0.08 |
ENST00000483339.2
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr9_-_215744 | 0.08 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr20_-_50418947 | 0.08 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_+_107224364 | 0.08 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr12_+_41136144 | 0.08 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr20_-_33735070 | 0.08 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr11_+_114168085 | 0.08 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr4_+_26344754 | 0.08 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_+_20081515 | 0.07 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chrX_-_71458802 | 0.07 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr2_-_136678123 | 0.07 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr3_-_185538849 | 0.07 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr10_-_75415825 | 0.07 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr1_+_160370344 | 0.07 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr4_+_87857538 | 0.07 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr2_+_202047843 | 0.07 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr7_-_87856280 | 0.07 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr12_+_28410128 | 0.07 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr15_+_93443419 | 0.07 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr18_-_51751132 | 0.07 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr7_-_87856303 | 0.07 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr12_+_64798826 | 0.07 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr4_-_170948361 | 0.07 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr19_+_50016610 | 0.07 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr13_-_81801115 | 0.06 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr8_+_132952112 | 0.06 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr4_-_185275104 | 0.06 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chrX_-_106243294 | 0.06 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr3_+_12329397 | 0.06 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr6_+_125304502 | 0.06 |
ENST00000519799.1
ENST00000368414.2 ENST00000359704.2 |
RNF217
|
ring finger protein 217 |
| chr17_+_48823896 | 0.06 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr17_-_64225508 | 0.06 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr17_-_45266542 | 0.06 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr2_-_191115229 | 0.06 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr1_+_12806141 | 0.06 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr20_+_22034809 | 0.06 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr2_-_231860596 | 0.06 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chrX_-_106243451 | 0.06 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr7_+_73868120 | 0.06 |
ENST00000265755.3
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr17_-_6524159 | 0.06 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr8_+_104384616 | 0.06 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr7_+_73868439 | 0.06 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr15_+_101417919 | 0.05 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr13_+_103459704 | 0.05 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr4_-_76944621 | 0.05 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr3_+_12329358 | 0.05 |
ENST00000309576.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr11_-_8857248 | 0.05 |
ENST00000534248.1
ENST00000530959.1 ENST00000531578.1 ENST00000533225.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr1_-_92371839 | 0.05 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr3_-_122233723 | 0.05 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr5_-_53115506 | 0.05 |
ENST00000511953.1
ENST00000504552.1 |
CTD-2081C10.1
|
CTD-2081C10.1 |
| chr14_-_21994337 | 0.05 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr1_-_92952433 | 0.05 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr2_+_27886330 | 0.05 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr2_-_8715616 | 0.05 |
ENST00000418358.1
|
AC011747.3
|
AC011747.3 |
| chr1_+_180601139 | 0.05 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr8_-_62602327 | 0.05 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr3_+_69812877 | 0.05 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr17_+_12569472 | 0.05 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr21_+_17909594 | 0.05 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr3_+_130569429 | 0.05 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr17_+_66511540 | 0.05 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr15_-_75748115 | 0.05 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr1_-_101360374 | 0.05 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr4_+_186990298 | 0.05 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr17_+_18086392 | 0.05 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr22_+_17956618 | 0.04 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr7_-_143599207 | 0.04 |
ENST00000355951.2
ENST00000479870.1 ENST00000478172.1 |
FAM115A
|
family with sequence similarity 115, member A |
| chr7_+_129932974 | 0.04 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr12_-_12674032 | 0.04 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr3_-_151034734 | 0.04 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr7_-_150020750 | 0.04 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr11_-_107729887 | 0.04 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_+_157154578 | 0.04 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr14_+_102276132 | 0.04 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_33686743 | 0.04 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr1_-_101360205 | 0.04 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr7_-_37488547 | 0.04 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_-_95487331 | 0.04 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr3_+_69812701 | 0.04 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr7_+_102553430 | 0.04 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr14_+_102276209 | 0.04 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr4_+_5527117 | 0.04 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr14_+_67831576 | 0.04 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr12_-_10282836 | 0.04 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr14_-_51027838 | 0.04 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_-_150020578 | 0.03 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr20_-_34117447 | 0.03 |
ENST00000246199.2
ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173
|
chromosome 20 open reading frame 173 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.4 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.1 | 0.7 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.3 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0010983 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 1.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.2 | GO:0072200 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:2000230 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 1.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.2 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.0 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0034699 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.2 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 1.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |