Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
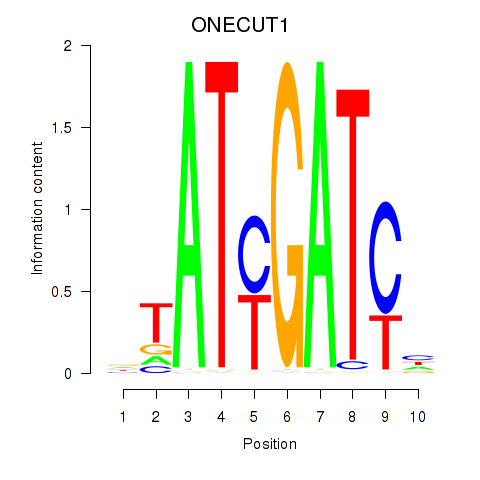
Results for ONECUT1
Z-value: 1.26
Transcription factors associated with ONECUT1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT1
|
ENSG00000169856.7 | one cut homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT1 | hg19_v2_chr15_-_53082178_53082209 | -0.65 | 1.6e-01 | Click! |
Activity profile of ONECUT1 motif
Sorted Z-values of ONECUT1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_42792442 | 2.61 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr11_+_57365150 | 0.77 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr6_+_27925019 | 0.73 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr1_-_186649543 | 0.61 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_-_145086922 | 0.58 |
ENST00000530478.1
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr18_+_47087390 | 0.50 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr13_-_86373536 | 0.48 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr14_+_24407940 | 0.47 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr4_-_157892055 | 0.47 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr4_-_111563076 | 0.45 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr17_-_56082455 | 0.45 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr2_-_192016276 | 0.45 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr8_-_23712312 | 0.43 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr5_+_32585605 | 0.41 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr1_-_238108575 | 0.41 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr14_+_38033252 | 0.41 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chrX_+_134654540 | 0.40 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr17_+_79369249 | 0.40 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr8_+_82192501 | 0.40 |
ENST00000297258.6
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated) |
| chr11_-_134123142 | 0.39 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chrM_+_14741 | 0.39 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr1_-_63782888 | 0.39 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr21_+_17961006 | 0.38 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_-_19729725 | 0.37 |
ENST00000251203.9
|
PBX4
|
pre-B-cell leukemia homeobox 4 |
| chr17_+_39868577 | 0.37 |
ENST00000329402.3
|
GAST
|
gastrin |
| chrM_+_8366 | 0.36 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr3_-_170744498 | 0.36 |
ENST00000382808.4
ENST00000314251.3 |
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr8_+_121457642 | 0.36 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr13_+_52586517 | 0.35 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr11_+_102188272 | 0.34 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr7_-_81635106 | 0.34 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr12_+_21679220 | 0.34 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr8_+_95558771 | 0.33 |
ENST00000391679.1
|
AC023632.1
|
HCG2009141; PRO2397; Uncharacterized protein |
| chr4_-_83483395 | 0.32 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr10_+_69869237 | 0.32 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr12_+_111471828 | 0.31 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr17_-_26697304 | 0.31 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr2_-_200715834 | 0.30 |
ENST00000420128.1
ENST00000416668.1 |
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr21_-_27107344 | 0.30 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr12_-_53297432 | 0.30 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr11_-_105948040 | 0.30 |
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr21_-_27107198 | 0.29 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr11_+_105948216 | 0.29 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr11_-_72385437 | 0.29 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr4_-_103940791 | 0.29 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr4_+_76649797 | 0.27 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr14_-_35873856 | 0.27 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr21_-_30445886 | 0.27 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr18_-_44775554 | 0.26 |
ENST00000425639.1
ENST00000400404.1 |
SKOR2
|
SKI family transcriptional corepressor 2 |
| chr9_-_77703115 | 0.26 |
ENST00000361092.4
ENST00000376808.4 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr3_+_107364769 | 0.26 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_+_198189921 | 0.26 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr9_-_2844058 | 0.26 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr4_+_79475019 | 0.26 |
ENST00000508214.1
|
ANXA3
|
annexin A3 |
| chr18_+_74207477 | 0.25 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr16_-_75467318 | 0.25 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr5_-_90679145 | 0.25 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr3_+_98699880 | 0.24 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr13_-_36920420 | 0.24 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr4_-_111563279 | 0.23 |
ENST00000511837.1
|
PITX2
|
paired-like homeodomain 2 |
| chr5_-_130868688 | 0.23 |
ENST00000504575.1
ENST00000513227.1 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr1_-_32384693 | 0.23 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr14_+_57671888 | 0.23 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr12_-_90024360 | 0.23 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_+_119799927 | 0.23 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr16_-_28192360 | 0.22 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr21_-_27107283 | 0.22 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr3_+_142315225 | 0.22 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr8_-_108510224 | 0.22 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr12_-_76817036 | 0.22 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr13_-_48612067 | 0.22 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr12_+_19389814 | 0.22 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr3_+_142315294 | 0.21 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr1_+_25598872 | 0.21 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr7_-_5998714 | 0.20 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr3_-_160117035 | 0.20 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr5_+_59726565 | 0.20 |
ENST00000412930.2
|
FKSG52
|
FKSG52 |
| chr22_+_51176624 | 0.20 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr11_+_47279155 | 0.20 |
ENST00000444396.1
ENST00000457932.1 ENST00000412937.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr3_+_69812877 | 0.20 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr1_-_1590418 | 0.20 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr11_-_10828892 | 0.20 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr6_-_131299929 | 0.20 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr4_+_159131596 | 0.19 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr4_-_141348999 | 0.19 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr4_+_107237660 | 0.19 |
ENST00000394701.4
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr19_-_13710678 | 0.19 |
ENST00000592864.1
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr3_+_191046810 | 0.18 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr12_-_13529594 | 0.18 |
ENST00000539026.1
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr3_-_160116995 | 0.18 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr3_+_107364683 | 0.18 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr18_-_68004529 | 0.18 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr19_-_58864848 | 0.18 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr5_-_75008244 | 0.17 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr15_-_64995399 | 0.17 |
ENST00000559753.1
ENST00000560258.2 ENST00000559912.2 ENST00000326005.6 |
OAZ2
|
ornithine decarboxylase antizyme 2 |
| chr12_-_56359802 | 0.17 |
ENST00000548803.1
ENST00000449260.2 ENST00000550447.1 ENST00000547137.1 ENST00000546543.1 ENST00000550464.1 |
PMEL
|
premelanosome protein |
| chr7_-_6523688 | 0.17 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr10_-_35104185 | 0.17 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr10_-_46167722 | 0.16 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr12_-_95941987 | 0.16 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chr7_-_154794763 | 0.16 |
ENST00000404141.1
|
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr20_+_22034809 | 0.16 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr20_-_58515344 | 0.16 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr18_+_68002675 | 0.16 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr11_-_105948129 | 0.16 |
ENST00000526793.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chrM_+_8527 | 0.16 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr16_-_49891694 | 0.16 |
ENST00000562520.1
|
ZNF423
|
zinc finger protein 423 |
| chr19_+_5681153 | 0.16 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr3_-_24207039 | 0.16 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr8_-_121457608 | 0.15 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr4_-_175443484 | 0.15 |
ENST00000514584.1
ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chrX_-_16887963 | 0.15 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr11_-_122930121 | 0.14 |
ENST00000524552.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr12_-_54653313 | 0.14 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr3_+_177545563 | 0.14 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr2_-_55646412 | 0.14 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr9_-_75653627 | 0.14 |
ENST00000446946.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr12_+_8995832 | 0.14 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr12_-_27924209 | 0.13 |
ENST00000381273.3
|
MANSC4
|
MANSC domain containing 4 |
| chr19_+_5681011 | 0.13 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr16_-_30621663 | 0.13 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr4_+_76649753 | 0.13 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr21_-_43816152 | 0.13 |
ENST00000433957.2
ENST00000398397.3 |
TMPRSS3
|
transmembrane protease, serine 3 |
| chr1_-_8075693 | 0.13 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr3_+_108308513 | 0.13 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr15_+_31196080 | 0.13 |
ENST00000561607.1
ENST00000565466.1 |
FAN1
|
FANCD2/FANCI-associated nuclease 1 |
| chr2_-_214017151 | 0.12 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_-_157892498 | 0.12 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chrM_-_14670 | 0.12 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr3_-_112693865 | 0.12 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr15_-_89089860 | 0.12 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr10_-_4720301 | 0.12 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr11_-_47788847 | 0.12 |
ENST00000263773.5
|
FNBP4
|
formin binding protein 4 |
| chr11_-_122931881 | 0.12 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr3_+_148415571 | 0.12 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr2_+_20650796 | 0.11 |
ENST00000448241.1
|
AC023137.2
|
AC023137.2 |
| chr1_-_165668100 | 0.11 |
ENST00000354775.4
|
ALDH9A1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr7_-_152373216 | 0.11 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr17_-_64225508 | 0.11 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr19_-_2721412 | 0.11 |
ENST00000323469.4
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr7_-_32529973 | 0.11 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_177939348 | 0.10 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr10_+_105881779 | 0.10 |
ENST00000369729.3
|
SFR1
|
SWI5-dependent recombination repair 1 |
| chr2_+_149804382 | 0.10 |
ENST00000397413.1
|
KIF5C
|
kinesin family member 5C |
| chr4_+_159131630 | 0.10 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr19_+_46800289 | 0.10 |
ENST00000377670.4
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr21_-_32716556 | 0.10 |
ENST00000455508.1
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr14_-_27066636 | 0.10 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr17_+_53342311 | 0.10 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr17_+_73539232 | 0.09 |
ENST00000580925.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr6_+_2246023 | 0.09 |
ENST00000530833.1
ENST00000525811.1 ENST00000534441.1 ENST00000533653.1 ENST00000534468.1 |
GMDS-AS1
|
GMDS antisense RNA 1 (head to head) |
| chr13_-_114018400 | 0.09 |
ENST00000375430.4
ENST00000375431.4 |
GRTP1
|
growth hormone regulated TBC protein 1 |
| chr19_+_13135386 | 0.09 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_+_129007964 | 0.09 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr5_+_167956121 | 0.09 |
ENST00000338333.4
|
FBLL1
|
fibrillarin-like 1 |
| chr10_+_51576285 | 0.09 |
ENST00000443446.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr3_+_188889737 | 0.09 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr10_+_105881906 | 0.08 |
ENST00000369727.3
ENST00000336358.5 |
SFR1
|
SWI5-dependent recombination repair 1 |
| chr7_-_69062391 | 0.08 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr16_-_75467274 | 0.08 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr2_+_157330081 | 0.08 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr6_-_31651817 | 0.08 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr13_-_46679185 | 0.08 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr4_-_83483360 | 0.08 |
ENST00000449862.2
|
TMEM150C
|
transmembrane protein 150C |
| chr6_+_126221034 | 0.08 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr10_+_28966271 | 0.08 |
ENST00000375533.3
|
BAMBI
|
BMP and activin membrane-bound inhibitor |
| chr15_+_38746307 | 0.08 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr6_+_108616243 | 0.08 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
| chr9_+_80850952 | 0.08 |
ENST00000424347.2
ENST00000415759.2 ENST00000376597.4 ENST00000277082.5 ENST00000376598.2 |
CEP78
|
centrosomal protein 78kDa |
| chr2_-_134326009 | 0.07 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr12_-_11422739 | 0.07 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr4_-_159644507 | 0.07 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr21_-_43816052 | 0.07 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr10_+_63661053 | 0.07 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr5_+_180794269 | 0.07 |
ENST00000456475.1
|
OR4F3
|
olfactory receptor, family 4, subfamily F, member 3 |
| chr7_-_26240357 | 0.07 |
ENST00000354667.4
ENST00000356674.7 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr21_+_27011899 | 0.07 |
ENST00000425221.2
|
JAM2
|
junctional adhesion molecule 2 |
| chr1_+_78245303 | 0.07 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr2_-_99870744 | 0.07 |
ENST00000409238.1
ENST00000423800.1 |
LYG2
|
lysozyme G-like 2 |
| chr13_+_52598827 | 0.07 |
ENST00000521776.2
|
UTP14C
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr14_+_74083548 | 0.06 |
ENST00000381139.1
|
ACOT6
|
acyl-CoA thioesterase 6 |
| chr2_-_39348137 | 0.06 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr8_+_22601 | 0.06 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr6_-_154568815 | 0.06 |
ENST00000519344.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chrX_+_10124977 | 0.06 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr3_+_101546827 | 0.06 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr12_-_8765446 | 0.05 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr12_-_96336369 | 0.05 |
ENST00000546947.1
ENST00000546386.1 |
CCDC38
|
coiled-coil domain containing 38 |
| chr5_-_60458179 | 0.05 |
ENST00000507416.1
ENST00000339020.3 |
SMIM15
|
small integral membrane protein 15 |
| chr12_-_13529642 | 0.05 |
ENST00000318426.2
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr1_-_165667545 | 0.05 |
ENST00000538148.1
|
ALDH9A1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr14_-_27066960 | 0.05 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr4_-_103747011 | 0.05 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_+_102980126 | 0.05 |
ENST00000375735.2
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr11_-_67407031 | 0.05 |
ENST00000335385.3
|
TBX10
|
T-box 10 |
| chr18_-_49557 | 0.05 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr11_+_107650219 | 0.05 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr3_+_108308559 | 0.05 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr22_+_24198890 | 0.05 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr6_-_27799305 | 0.05 |
ENST00000357549.2
|
HIST1H4K
|
histone cluster 1, H4k |
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.7 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.1 | 2.6 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 0.9 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.2 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 0.3 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0007621 | courtship behavior(GO:0007619) negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.2 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.3 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0060717 | chorion development(GO:0060717) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.1 | 0.3 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 1.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |