Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
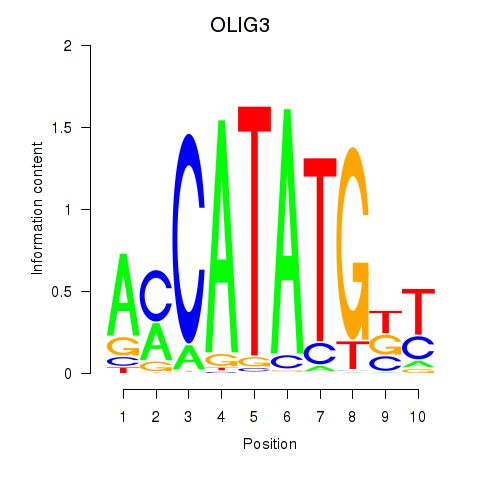
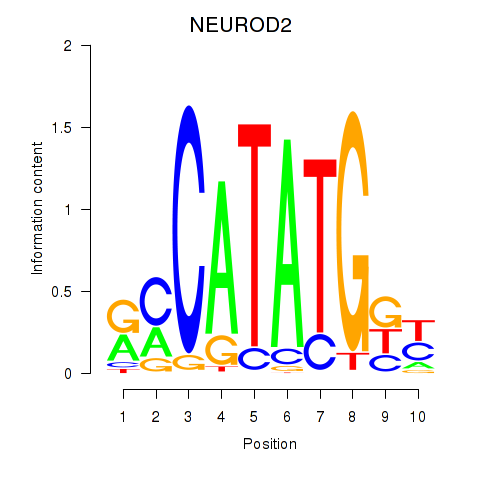
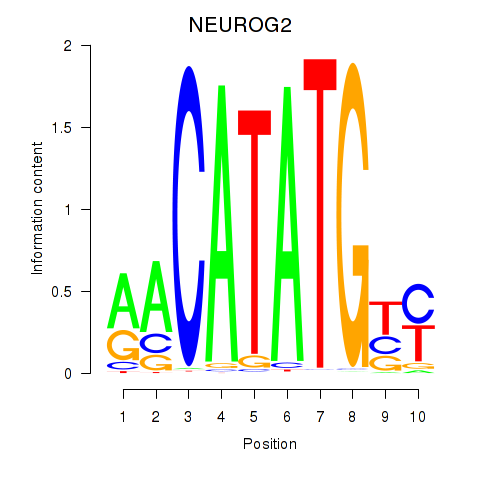
Results for OLIG3_NEUROD2_NEUROG2
Z-value: 0.52
Transcription factors associated with OLIG3_NEUROD2_NEUROG2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG3
|
ENSG00000177468.5 | oligodendrocyte transcription factor 3 |
|
NEUROD2
|
ENSG00000171532.4 | neuronal differentiation 2 |
|
NEUROG2
|
ENSG00000178403.3 | neurogenin 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NEUROD2 | hg19_v2_chr17_-_37764128_37764258 | 0.31 | 5.5e-01 | Click! |
Activity profile of OLIG3_NEUROD2_NEUROG2 motif
Sorted Z-values of OLIG3_NEUROD2_NEUROG2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_2912363 | 1.06 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr10_+_89124746 | 0.54 |
ENST00000465545.1
|
NUTM2D
|
NUT family member 2D |
| chr8_-_112248400 | 0.47 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr2_+_105471969 | 0.36 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr3_-_197300194 | 0.35 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr15_-_98417780 | 0.35 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr19_-_6057282 | 0.34 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr21_+_38593701 | 0.33 |
ENST00000440629.1
|
AP001432.14
|
AP001432.14 |
| chr5_+_67535647 | 0.27 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr17_-_65992544 | 0.27 |
ENST00000580729.1
|
RP11-855A2.5
|
RP11-855A2.5 |
| chr1_+_81106951 | 0.26 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr1_+_196743943 | 0.24 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr6_+_155585147 | 0.24 |
ENST00000367165.3
|
CLDN20
|
claudin 20 |
| chr7_-_48068643 | 0.24 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr1_-_114429997 | 0.21 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr1_+_95975672 | 0.21 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr14_+_32414059 | 0.20 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr12_+_79371565 | 0.20 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr4_+_174818390 | 0.20 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr11_-_26743546 | 0.19 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr1_-_168464875 | 0.19 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr22_+_20877924 | 0.19 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr4_-_111563076 | 0.19 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr16_+_67280799 | 0.17 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr2_-_132589601 | 0.17 |
ENST00000437330.1
|
AC103564.7
|
AC103564.7 |
| chr17_+_73629500 | 0.17 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr10_-_69597828 | 0.16 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr2_+_177053307 | 0.16 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr8_+_101349823 | 0.16 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr3_+_185300270 | 0.16 |
ENST00000430355.1
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr14_+_32476072 | 0.16 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr4_+_144354644 | 0.16 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr3_-_52486841 | 0.15 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr12_-_11036844 | 0.15 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr1_-_186344802 | 0.15 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr12_-_123450986 | 0.15 |
ENST00000344275.7
ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr6_-_31648150 | 0.14 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr10_-_9801179 | 0.14 |
ENST00000419836.1
|
RP5-1051H14.2
|
RP5-1051H14.2 |
| chrX_+_70798261 | 0.14 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr6_-_150219232 | 0.14 |
ENST00000531073.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr16_+_53133070 | 0.14 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr2_-_105953912 | 0.14 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr15_-_38519066 | 0.13 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr7_-_24957699 | 0.13 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr17_+_68047418 | 0.12 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr10_+_6392278 | 0.12 |
ENST00000391437.1
|
DKFZP667F0711
|
DKFZP667F0711 |
| chr3_+_72201910 | 0.12 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr4_+_25915896 | 0.12 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr2_-_863877 | 0.12 |
ENST00000415700.1
|
AC113607.1
|
long intergenic non-protein coding RNA 1115 |
| chr14_-_107283278 | 0.12 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr8_-_41508855 | 0.12 |
ENST00000518699.2
|
NKX6-3
|
NK6 homeobox 3 |
| chr16_-_85969774 | 0.12 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr12_+_133656995 | 0.12 |
ENST00000356456.5
|
ZNF140
|
zinc finger protein 140 |
| chr10_-_69597810 | 0.12 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr14_+_95047744 | 0.11 |
ENST00000553511.1
ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr7_-_101212244 | 0.11 |
ENST00000451953.1
ENST00000434537.1 ENST00000437900.1 |
LINC01007
|
long intergenic non-protein coding RNA 1007 |
| chr6_-_56716686 | 0.11 |
ENST00000520645.1
|
DST
|
dystonin |
| chr4_+_160188889 | 0.11 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr4_-_171011084 | 0.11 |
ENST00000337664.4
|
AADAT
|
aminoadipate aminotransferase |
| chr3_-_52719546 | 0.11 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr3_-_195310802 | 0.10 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr10_+_85954377 | 0.10 |
ENST00000332904.3
ENST00000372117.3 |
CDHR1
|
cadherin-related family member 1 |
| chr9_-_95244781 | 0.10 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr7_+_141478242 | 0.10 |
ENST00000247881.2
|
TAS2R4
|
taste receptor, type 2, member 4 |
| chr10_+_44101850 | 0.10 |
ENST00000361807.3
ENST00000374437.2 ENST00000430885.1 ENST00000374435.3 |
ZNF485
|
zinc finger protein 485 |
| chr7_+_114562909 | 0.10 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr16_-_21877629 | 0.10 |
ENST00000544693.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr12_-_106697974 | 0.10 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr18_+_59415396 | 0.10 |
ENST00000567801.1
|
RP11-1096D5.1
|
RP11-1096D5.1 |
| chr1_+_180875711 | 0.10 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr12_-_123634449 | 0.10 |
ENST00000542210.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr11_-_62457371 | 0.10 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr16_-_14726576 | 0.09 |
ENST00000562896.1
|
PARN
|
poly(A)-specific ribonuclease |
| chr3_+_112930387 | 0.09 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr1_+_110881945 | 0.09 |
ENST00000602849.1
ENST00000487146.2 |
RBM15
|
RNA binding motif protein 15 |
| chr3_+_113667354 | 0.09 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr7_-_55930443 | 0.09 |
ENST00000388975.3
|
SEPT14
|
septin 14 |
| chr1_-_169764026 | 0.09 |
ENST00000454472.1
ENST00000310392.4 |
METTL18
|
methyltransferase like 18 |
| chr11_-_4629388 | 0.09 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr3_+_113251143 | 0.09 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr15_+_38964048 | 0.09 |
ENST00000560203.1
ENST00000557946.1 |
RP11-275I4.2
|
RP11-275I4.2 |
| chr4_+_114214125 | 0.09 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr3_-_114477962 | 0.09 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr19_-_21512115 | 0.09 |
ENST00000601295.1
|
ZNF708
|
zinc finger protein 708 |
| chr4_-_156297919 | 0.08 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr2_-_158345462 | 0.08 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr4_+_53525573 | 0.08 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr1_-_33116128 | 0.08 |
ENST00000436661.1
ENST00000373501.2 ENST00000341885.5 ENST00000468695.1 |
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand |
| chr16_-_72698834 | 0.08 |
ENST00000570152.1
ENST00000561611.2 ENST00000570035.1 |
AC004158.2
|
AC004158.2 |
| chr5_-_158757895 | 0.08 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr17_+_48046671 | 0.08 |
ENST00000505318.2
|
DLX4
|
distal-less homeobox 4 |
| chr3_-_114477787 | 0.08 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_-_134123142 | 0.08 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chrX_+_69509870 | 0.08 |
ENST00000374388.3
|
KIF4A
|
kinesin family member 4A |
| chr12_+_58335360 | 0.08 |
ENST00000300145.3
|
XRCC6BP1
|
XRCC6 binding protein 1 |
| chr6_+_3000195 | 0.08 |
ENST00000338130.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr4_+_2813946 | 0.07 |
ENST00000442312.2
|
SH3BP2
|
SH3-domain binding protein 2 |
| chrX_-_13752675 | 0.07 |
ENST00000380579.1
ENST00000458511.2 ENST00000519885.1 ENST00000358231.5 ENST00000518847.1 ENST00000453655.2 ENST00000359680.5 |
TRAPPC2
|
trafficking protein particle complex 2 |
| chr16_-_28550348 | 0.07 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr14_+_69865401 | 0.07 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr10_-_15902449 | 0.07 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chrX_+_118108601 | 0.07 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr1_+_104293028 | 0.07 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr6_+_3000057 | 0.07 |
ENST00000397717.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr14_+_52164675 | 0.07 |
ENST00000555936.1
|
FRMD6
|
FERM domain containing 6 |
| chr2_+_54342574 | 0.07 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr5_-_137674000 | 0.07 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr3_+_187420101 | 0.07 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr16_-_18887627 | 0.07 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr2_-_70409953 | 0.07 |
ENST00000419381.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr18_-_10748498 | 0.07 |
ENST00000579949.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr4_-_24914508 | 0.07 |
ENST00000504487.1
|
CCDC149
|
coiled-coil domain containing 149 |
| chr17_-_191188 | 0.07 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr8_+_39972170 | 0.07 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr11_+_6226782 | 0.07 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr4_-_146019335 | 0.07 |
ENST00000451299.2
ENST00000507656.1 ENST00000309439.5 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr7_+_105172612 | 0.06 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr11_+_7595136 | 0.06 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr8_-_133772870 | 0.06 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr18_-_74839891 | 0.06 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr4_+_187065978 | 0.06 |
ENST00000227065.4
ENST00000502970.1 ENST00000514153.1 |
FAM149A
|
family with sequence similarity 149, member A |
| chr16_-_55867146 | 0.06 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr3_-_178984759 | 0.06 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chrX_+_2670066 | 0.06 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr14_+_83108955 | 0.06 |
ENST00000555798.1
ENST00000553760.1 ENST00000555150.1 ENST00000556970.1 |
RP11-406A9.2
|
RP11-406A9.2 |
| chr10_+_133918175 | 0.06 |
ENST00000298622.4
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr1_+_156182773 | 0.06 |
ENST00000490491.1
ENST00000368276.4 ENST00000320139.5 ENST00000368279.3 ENST00000368273.4 ENST00000368277.3 ENST00000567140.1 ENST00000565805.1 |
PMF1-BGLAP
PMF1
|
PMF1-BGLAP readthrough polyamine-modulated factor 1 |
| chr1_+_89990378 | 0.06 |
ENST00000449440.1
|
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr17_+_29898161 | 0.06 |
ENST00000412403.1
|
AC003101.1
|
Uncharacterized protein |
| chrX_+_69509927 | 0.06 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr15_-_55700522 | 0.06 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chrX_+_37765364 | 0.06 |
ENST00000567273.1
|
AL121578.2
|
AL121578.2 |
| chr17_-_39538550 | 0.06 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr13_+_77564795 | 0.06 |
ENST00000377453.3
|
CLN5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr19_+_40005753 | 0.06 |
ENST00000335426.4
ENST00000423711.1 |
SELV
|
Selenoprotein V |
| chr1_-_114430169 | 0.06 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr10_-_118429461 | 0.06 |
ENST00000588184.1
ENST00000369210.3 |
C10orf82
|
chromosome 10 open reading frame 82 |
| chrX_-_102565932 | 0.06 |
ENST00000372674.1
ENST00000372677.3 |
BEX2
|
brain expressed X-linked 2 |
| chr15_-_45670924 | 0.06 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr15_-_65503801 | 0.06 |
ENST00000261883.4
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr5_+_175299743 | 0.05 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr15_-_80189380 | 0.05 |
ENST00000258874.3
|
MTHFS
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr12_+_54393880 | 0.05 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chrX_+_47444613 | 0.05 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr1_+_87458692 | 0.05 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr1_-_144994840 | 0.05 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr10_-_69597915 | 0.05 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_-_220220000 | 0.05 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr19_-_58204128 | 0.05 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr4_+_175205162 | 0.05 |
ENST00000503053.1
|
CEP44
|
centrosomal protein 44kDa |
| chrX_-_102757802 | 0.05 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr6_+_2988199 | 0.05 |
ENST00000450238.1
ENST00000445000.1 ENST00000426637.1 |
LINC01011
NQO2
|
long intergenic non-protein coding RNA 1011 NAD(P)H dehydrogenase, quinone 2 |
| chr21_+_46654249 | 0.05 |
ENST00000584169.1
ENST00000328344.2 |
LINC00334
|
long intergenic non-protein coding RNA 334 |
| chr4_+_57276661 | 0.05 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr3_-_105588231 | 0.05 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr17_-_15175604 | 0.05 |
ENST00000431343.1
|
AC005703.3
|
AC005703.3 |
| chr9_-_127703333 | 0.05 |
ENST00000373555.4
|
GOLGA1
|
golgin A1 |
| chr1_+_205538105 | 0.05 |
ENST00000367147.4
ENST00000539267.1 |
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr12_-_67197760 | 0.05 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr11_-_8795787 | 0.05 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr10_-_123357598 | 0.05 |
ENST00000358487.5
ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr10_-_75415825 | 0.05 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr12_-_11002063 | 0.05 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr11_-_11374904 | 0.05 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr18_+_13465008 | 0.05 |
ENST00000593236.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr5_-_114632307 | 0.05 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr1_-_45272951 | 0.05 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr8_-_16859690 | 0.05 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr5_-_159766528 | 0.05 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr15_+_91418918 | 0.05 |
ENST00000560824.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr10_-_94257512 | 0.05 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr14_-_75735986 | 0.05 |
ENST00000553510.1
|
RP11-293M10.1
|
Uncharacterized protein |
| chr10_-_52000307 | 0.05 |
ENST00000443575.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr2_-_8715616 | 0.05 |
ENST00000418358.1
|
AC011747.3
|
AC011747.3 |
| chr17_-_72619869 | 0.05 |
ENST00000392619.1
ENST00000426295.2 |
CD300E
|
CD300e molecule |
| chr5_-_60140089 | 0.05 |
ENST00000507047.1
ENST00000438340.1 ENST00000425382.1 ENST00000508821.1 |
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr8_-_23315131 | 0.05 |
ENST00000518718.1
|
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr5_-_135701164 | 0.05 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr8_+_67782984 | 0.05 |
ENST00000396592.3
ENST00000422365.2 ENST00000492775.1 |
MCMDC2
|
minichromosome maintenance domain containing 2 |
| chr10_+_99627889 | 0.05 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr1_-_223853348 | 0.05 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr4_+_175205100 | 0.05 |
ENST00000515299.1
|
CEP44
|
centrosomal protein 44kDa |
| chr9_-_37904084 | 0.05 |
ENST00000377716.2
ENST00000242275.6 |
SLC25A51
|
solute carrier family 25, member 51 |
| chr6_-_27880174 | 0.05 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr11_-_795170 | 0.05 |
ENST00000481290.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr11_-_6633799 | 0.05 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr1_-_220219775 | 0.05 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr9_+_135037334 | 0.05 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr12_+_10124001 | 0.05 |
ENST00000396507.3
ENST00000304361.4 ENST00000434319.2 |
CLEC12A
|
C-type lectin domain family 12, member A |
| chr4_-_24914576 | 0.05 |
ENST00000502801.1
ENST00000428116.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr4_+_47487285 | 0.04 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr11_-_96239990 | 0.04 |
ENST00000511243.2
|
JRKL-AS1
|
JRKL antisense RNA 1 |
| chr3_+_179040767 | 0.04 |
ENST00000496856.1
ENST00000491818.1 |
ZNF639
|
zinc finger protein 639 |
| chr9_-_73029540 | 0.04 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr14_-_75079294 | 0.04 |
ENST00000556359.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr2_+_11674213 | 0.04 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr14_+_95047725 | 0.04 |
ENST00000554760.1
ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr12_+_94551874 | 0.04 |
ENST00000551850.1
|
PLXNC1
|
plexin C1 |
| chr1_+_144215387 | 0.04 |
ENST00000421555.2
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr16_+_28505955 | 0.04 |
ENST00000564831.1
ENST00000328423.5 ENST00000431282.1 |
APOBR
|
apolipoprotein B receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG3_NEUROD2_NEUROG2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072069 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 1.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0033076 | isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.1 | GO:0060667 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.0 | 0.0 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.0 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097180 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.2 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |