Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
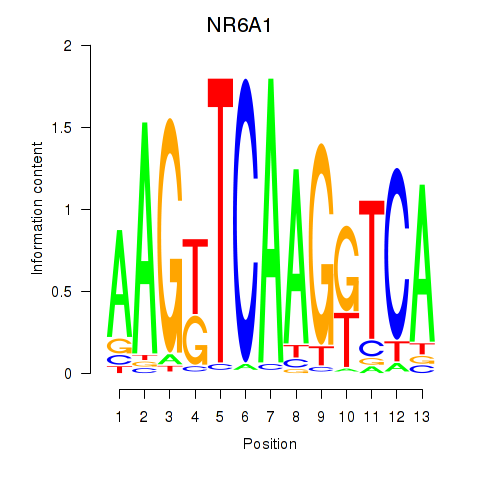
Results for NR6A1
Z-value: 0.40
Transcription factors associated with NR6A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR6A1
|
ENSG00000148200.12 | nuclear receptor subfamily 6 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR6A1 | hg19_v2_chr9_-_127358087_127358170 | 0.60 | 2.1e-01 | Click! |
Activity profile of NR6A1 motif
Sorted Z-values of NR6A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_185776854 | 0.33 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr22_-_28490123 | 0.31 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr12_+_54384370 | 0.31 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr19_+_11670245 | 0.27 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr11_+_115498761 | 0.26 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr16_-_21875424 | 0.21 |
ENST00000541674.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr19_-_6057282 | 0.21 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr22_+_45680822 | 0.20 |
ENST00000216211.4
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr19_-_51289374 | 0.20 |
ENST00000563228.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr1_-_54879140 | 0.19 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr11_-_72852320 | 0.19 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr11_-_104480019 | 0.19 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr16_-_30997533 | 0.17 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr2_+_33701684 | 0.17 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chrX_+_48620147 | 0.16 |
ENST00000303227.6
|
GLOD5
|
glyoxalase domain containing 5 |
| chr11_-_33743952 | 0.16 |
ENST00000534312.1
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr16_+_777739 | 0.16 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr12_+_2921788 | 0.15 |
ENST00000228799.2
ENST00000419778.2 ENST00000542548.1 |
ITFG2
|
integrin alpha FG-GAP repeat containing 2 |
| chr17_+_44679808 | 0.15 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr14_+_76071805 | 0.15 |
ENST00000539311.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr7_-_151107767 | 0.14 |
ENST00000477459.1
|
WDR86
|
WD repeat domain 86 |
| chr7_-_150038704 | 0.14 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr10_-_14613968 | 0.14 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr12_-_24737089 | 0.14 |
ENST00000483544.1
|
LINC00477
|
long intergenic non-protein coding RNA 477 |
| chr19_+_41620335 | 0.14 |
ENST00000331105.2
|
CYP2F1
|
cytochrome P450, family 2, subfamily F, polypeptide 1 |
| chr7_-_69062391 | 0.14 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr2_-_178128149 | 0.13 |
ENST00000423513.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr16_+_20817746 | 0.13 |
ENST00000568894.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr15_+_41057818 | 0.13 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr21_-_32716556 | 0.13 |
ENST00000455508.1
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr19_-_43709772 | 0.13 |
ENST00000596907.1
ENST00000451895.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr7_-_56160625 | 0.13 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr6_+_5085572 | 0.13 |
ENST00000405617.2
|
PPP1R3G
|
protein phosphatase 1, regulatory subunit 3G |
| chr2_-_14541060 | 0.13 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr9_+_132096166 | 0.12 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr14_+_89029323 | 0.12 |
ENST00000554602.1
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr10_+_695888 | 0.12 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr2_+_175352114 | 0.12 |
ENST00000444196.1
ENST00000417038.1 ENST00000606406.1 |
AC010894.3
|
AC010894.3 |
| chr4_+_57253672 | 0.12 |
ENST00000602927.1
|
RP11-646I6.5
|
RP11-646I6.5 |
| chr12_+_26164645 | 0.12 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr4_-_170948361 | 0.12 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr5_-_55902050 | 0.12 |
ENST00000438651.1
|
AC022431.2
|
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
| chr17_-_18945798 | 0.12 |
ENST00000395635.1
|
GRAP
|
GRB2-related adaptor protein |
| chr12_+_57810198 | 0.12 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr17_+_55173955 | 0.11 |
ENST00000576591.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr12_+_69186125 | 0.11 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr19_-_16008880 | 0.11 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr19_-_22193731 | 0.11 |
ENST00000601773.1
ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208
|
zinc finger protein 208 |
| chr7_+_99425633 | 0.11 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr14_+_59100774 | 0.11 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr5_+_140749803 | 0.11 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr11_-_64014379 | 0.11 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr4_-_10459009 | 0.11 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr12_+_116997186 | 0.11 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr3_+_171561127 | 0.11 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr9_-_14314518 | 0.10 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr20_-_62738524 | 0.10 |
ENST00000369768.1
|
NPBWR2
|
neuropeptides B/W receptor 2 |
| chr10_-_14614095 | 0.10 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr19_-_10446449 | 0.10 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr5_+_55147205 | 0.09 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr2_-_211090162 | 0.09 |
ENST00000233710.3
|
ACADL
|
acyl-CoA dehydrogenase, long chain |
| chr7_+_95171457 | 0.09 |
ENST00000601424.1
|
AC002451.1
|
Protein LOC100996577 |
| chr19_-_8373173 | 0.09 |
ENST00000537716.2
ENST00000301458.5 |
CD320
|
CD320 molecule |
| chrY_+_16636354 | 0.09 |
ENST00000339174.5
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr16_+_31085714 | 0.08 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr4_+_159593271 | 0.08 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr19_-_43244610 | 0.08 |
ENST00000595140.1
ENST00000327495.5 |
PSG3
|
pregnancy specific beta-1-glycoprotein 3 |
| chr10_-_14614311 | 0.08 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr6_+_30856507 | 0.08 |
ENST00000513240.1
ENST00000424544.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_+_113185292 | 0.08 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chr8_-_145701718 | 0.08 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr11_-_128775592 | 0.08 |
ENST00000310799.3
|
C11orf45
|
chromosome 11 open reading frame 45 |
| chrX_+_22050546 | 0.08 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr3_+_57882024 | 0.08 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr19_-_22193706 | 0.08 |
ENST00000597040.1
|
ZNF208
|
zinc finger protein 208 |
| chr19_+_42381337 | 0.08 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr4_+_172734548 | 0.08 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr19_+_17516909 | 0.08 |
ENST00000601007.1
ENST00000594913.1 ENST00000599975.1 ENST00000600514.1 |
CTD-2521M24.9
MVB12A
|
CTD-2521M24.9 multivesicular body subunit 12A |
| chr18_+_11751493 | 0.08 |
ENST00000269162.5
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr11_-_33744256 | 0.08 |
ENST00000415002.2
ENST00000437761.2 ENST00000445143.2 |
CD59
|
CD59 molecule, complement regulatory protein |
| chr18_+_11751466 | 0.07 |
ENST00000535121.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr8_+_9046503 | 0.07 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr17_-_45056606 | 0.07 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr5_-_59064458 | 0.07 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_1248547 | 0.07 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr19_-_43690674 | 0.07 |
ENST00000342951.6
ENST00000366175.3 |
PSG5
|
pregnancy specific beta-1-glycoprotein 5 |
| chrX_-_118827113 | 0.07 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr6_-_31620095 | 0.07 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr6_+_25652501 | 0.07 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr15_+_64752927 | 0.07 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chr16_+_67195592 | 0.07 |
ENST00000519378.1
|
FBXL8
|
F-box and leucine-rich repeat protein 8 |
| chr3_+_32993065 | 0.07 |
ENST00000330953.5
|
CCR4
|
chemokine (C-C motif) receptor 4 |
| chr19_+_42381173 | 0.07 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr16_-_11723066 | 0.06 |
ENST00000576036.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr11_-_45940343 | 0.06 |
ENST00000532681.1
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr1_+_6684918 | 0.06 |
ENST00000054650.4
|
THAP3
|
THAP domain containing, apoptosis associated protein 3 |
| chr12_+_54410664 | 0.06 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr13_-_95131923 | 0.06 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr10_-_14614122 | 0.06 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr10_+_47894023 | 0.06 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr10_-_76995675 | 0.06 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr1_-_91813006 | 0.06 |
ENST00000430465.1
|
HFM1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr7_+_94536898 | 0.06 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr3_+_185046676 | 0.06 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr9_+_33795533 | 0.06 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr7_+_142985467 | 0.06 |
ENST00000392925.2
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr5_-_14871866 | 0.06 |
ENST00000284268.6
|
ANKH
|
ANKH inorganic pyrophosphate transport regulator |
| chr1_+_45212074 | 0.06 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr12_-_10320256 | 0.06 |
ENST00000538745.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr12_-_48214345 | 0.06 |
ENST00000433685.1
ENST00000447463.1 ENST00000427332.2 |
HDAC7
|
histone deacetylase 7 |
| chr12_+_108956355 | 0.06 |
ENST00000535729.1
ENST00000431221.2 ENST00000547005.1 ENST00000311893.9 ENST00000392807.4 ENST00000338291.4 ENST00000539593.1 |
ISCU
|
iron-sulfur cluster assembly enzyme |
| chr14_-_20020272 | 0.06 |
ENST00000551509.1
|
POTEM
|
POTE ankyrin domain family, member M |
| chr9_-_130679257 | 0.06 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr1_-_183538319 | 0.06 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr16_+_69166770 | 0.06 |
ENST00000567235.2
ENST00000568448.1 |
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr4_+_668348 | 0.06 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr6_+_43028182 | 0.06 |
ENST00000394058.1
|
KLC4
|
kinesin light chain 4 |
| chr3_-_126327398 | 0.06 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr3_-_52312337 | 0.06 |
ENST00000469000.1
|
WDR82
|
WD repeat domain 82 |
| chr6_+_30851205 | 0.05 |
ENST00000515881.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chrX_+_48916497 | 0.05 |
ENST00000496529.2
ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120
|
coiled-coil domain containing 120 |
| chr10_+_123872483 | 0.05 |
ENST00000369001.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_+_99933730 | 0.05 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr5_+_140579162 | 0.05 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr1_-_228594490 | 0.05 |
ENST00000366699.3
ENST00000284551.6 |
TRIM11
|
tripartite motif containing 11 |
| chr5_-_140027357 | 0.05 |
ENST00000252102.4
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr6_-_43027105 | 0.05 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr5_-_138725560 | 0.05 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr19_+_49956426 | 0.05 |
ENST00000293350.4
ENST00000540132.1 ENST00000455361.2 ENST00000433981.2 |
ALDH16A1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr3_+_113465866 | 0.05 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr9_-_97090926 | 0.05 |
ENST00000335456.7
ENST00000253262.4 ENST00000341207.4 |
NUTM2F
|
NUT family member 2F |
| chr16_-_18801582 | 0.05 |
ENST00000565420.1
|
RPS15A
|
ribosomal protein S15a |
| chr17_+_47448102 | 0.05 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr10_-_120101752 | 0.05 |
ENST00000369170.4
|
FAM204A
|
family with sequence similarity 204, member A |
| chr2_-_86790472 | 0.05 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chr9_-_14313641 | 0.05 |
ENST00000380953.1
|
NFIB
|
nuclear factor I/B |
| chr12_-_7261772 | 0.05 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr7_+_99202003 | 0.05 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr17_+_48503519 | 0.05 |
ENST00000300441.4
ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr17_-_7531121 | 0.05 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr1_+_145289772 | 0.05 |
ENST00000369339.3
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr2_+_101179152 | 0.05 |
ENST00000264254.6
|
PDCL3
|
phosducin-like 3 |
| chr6_+_30844192 | 0.05 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_-_108464321 | 0.05 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr9_+_15553055 | 0.05 |
ENST00000380701.3
|
CCDC171
|
coiled-coil domain containing 171 |
| chr10_+_106014468 | 0.05 |
ENST00000369710.4
ENST00000369713.5 ENST00000445155.1 |
GSTO1
|
glutathione S-transferase omega 1 |
| chr6_+_28109703 | 0.05 |
ENST00000457389.2
ENST00000330236.6 |
ZKSCAN8
|
zinc finger with KRAB and SCAN domains 8 |
| chr12_+_58087738 | 0.05 |
ENST00000552285.1
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr1_+_45212051 | 0.05 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr13_-_19755975 | 0.05 |
ENST00000400113.3
|
TUBA3C
|
tubulin, alpha 3c |
| chr11_-_64512273 | 0.04 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr2_+_219575653 | 0.04 |
ENST00000442769.1
ENST00000424644.1 |
TTLL4
|
tubulin tyrosine ligase-like family, member 4 |
| chr1_-_207095212 | 0.04 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr11_+_62652649 | 0.04 |
ENST00000539507.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr1_-_146082633 | 0.04 |
ENST00000605317.1
ENST00000604938.1 ENST00000339388.5 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr13_-_24463530 | 0.04 |
ENST00000382172.3
|
MIPEP
|
mitochondrial intermediate peptidase |
| chrX_+_47229982 | 0.04 |
ENST00000377073.3
|
ZNF157
|
zinc finger protein 157 |
| chr9_-_38069208 | 0.04 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr2_-_26467557 | 0.04 |
ENST00000380649.3
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr2_+_201994208 | 0.04 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_57881966 | 0.04 |
ENST00000495364.1
|
SLMAP
|
sarcolemma associated protein |
| chr14_+_19553365 | 0.04 |
ENST00000409832.3
|
POTEG
|
POTE ankyrin domain family, member G |
| chr12_+_122242597 | 0.04 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr5_-_140700322 | 0.04 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr9_-_39239171 | 0.04 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr7_-_642261 | 0.04 |
ENST00000400758.2
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr13_+_111766897 | 0.04 |
ENST00000317133.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr12_+_57623477 | 0.04 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chrX_-_118827333 | 0.04 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr11_-_68518910 | 0.04 |
ENST00000544963.1
ENST00000443940.2 |
MTL5
|
metallothionein-like 5, testis-specific (tesmin) |
| chr6_+_106534192 | 0.04 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr2_-_207024233 | 0.04 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr19_-_58864848 | 0.04 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr11_+_113185533 | 0.04 |
ENST00000393020.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr11_-_78052923 | 0.04 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr16_-_12897642 | 0.04 |
ENST00000433677.2
ENST00000261660.4 ENST00000381774.4 |
CPPED1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr6_+_25652432 | 0.04 |
ENST00000377961.2
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr19_+_7985198 | 0.04 |
ENST00000221573.6
ENST00000595637.1 |
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr4_+_7045042 | 0.04 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
| chr8_-_38126675 | 0.04 |
ENST00000531823.1
ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr1_-_20126365 | 0.04 |
ENST00000294543.6
ENST00000375122.2 |
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr19_-_49956728 | 0.04 |
ENST00000601825.1
ENST00000596049.1 ENST00000599366.1 ENST00000597415.1 |
PIH1D1
|
PIH1 domain containing 1 |
| chr17_-_37308824 | 0.03 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr2_+_219283815 | 0.03 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr5_+_140027355 | 0.03 |
ENST00000417647.2
ENST00000507593.1 ENST00000508301.1 |
IK
|
IK cytokine, down-regulator of HLA II |
| chr10_-_126107482 | 0.03 |
ENST00000368845.5
ENST00000539214.1 |
OAT
|
ornithine aminotransferase |
| chr10_+_106013923 | 0.03 |
ENST00000539281.1
|
GSTO1
|
glutathione S-transferase omega 1 |
| chr10_+_46222648 | 0.03 |
ENST00000336378.4
ENST00000540872.1 ENST00000537517.1 ENST00000374362.2 ENST00000359860.4 ENST00000420848.1 |
FAM21C
|
family with sequence similarity 21, member C |
| chr10_-_70092671 | 0.03 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr20_+_43803517 | 0.03 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr2_-_26467465 | 0.03 |
ENST00000457468.2
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr3_-_139108475 | 0.03 |
ENST00000515006.1
ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr3_+_14716606 | 0.03 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr2_-_86790593 | 0.03 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr4_+_185973026 | 0.03 |
ENST00000506338.1
ENST00000509017.1 |
RP11-386B13.3
|
RP11-386B13.3 |
| chr10_-_75634219 | 0.03 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr8_+_144640477 | 0.03 |
ENST00000262580.4
|
GSDMD
|
gasdermin D |
| chr10_-_14574705 | 0.03 |
ENST00000489100.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr9_+_131217459 | 0.03 |
ENST00000497812.2
ENST00000393533.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr22_-_45609315 | 0.03 |
ENST00000443310.3
ENST00000424508.1 |
KIAA0930
|
KIAA0930 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR6A1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.2 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.0 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |