Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
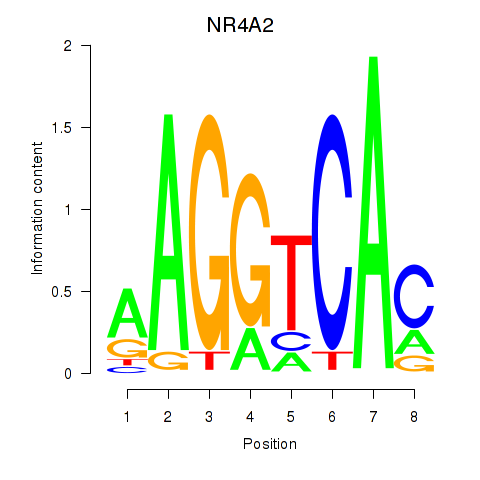
Results for NR4A2
Z-value: 0.58
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.9 | nuclear receptor subfamily 4 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A2 | hg19_v2_chr2_-_157189180_157189290 | -0.54 | 2.7e-01 | Click! |
Activity profile of NR4A2 motif
Sorted Z-values of NR4A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_228678550 | 0.91 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr14_+_21492331 | 0.62 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr11_-_790060 | 0.55 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr1_+_45140400 | 0.47 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr15_-_42565023 | 0.43 |
ENST00000566474.1
|
TMEM87A
|
transmembrane protein 87A |
| chr3_-_113464906 | 0.41 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr8_-_131028869 | 0.39 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr10_-_94301107 | 0.39 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr1_+_196621002 | 0.35 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr17_-_4889508 | 0.32 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr1_+_196621156 | 0.32 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr3_-_167452298 | 0.31 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr11_+_76571911 | 0.30 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr22_-_32341336 | 0.30 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr8_-_131028641 | 0.29 |
ENST00000523509.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr11_-_47470591 | 0.29 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chrY_-_15591818 | 0.28 |
ENST00000382893.1
|
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr4_-_71705082 | 0.26 |
ENST00000439371.1
ENST00000499044.2 |
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr8_-_53626974 | 0.26 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr20_-_13765526 | 0.26 |
ENST00000202816.1
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chrX_+_106045891 | 0.25 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr3_-_167452262 | 0.25 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr11_+_86748863 | 0.25 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr15_+_43885252 | 0.25 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr12_-_16760021 | 0.24 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr10_+_22605374 | 0.24 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr2_+_162016827 | 0.24 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr3_+_179322481 | 0.24 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr21_-_35014027 | 0.23 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr2_+_162016804 | 0.22 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr11_+_86749035 | 0.22 |
ENST00000305494.5
ENST00000535167.1 |
TMEM135
|
transmembrane protein 135 |
| chr1_-_197169672 | 0.22 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr4_-_71705027 | 0.22 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr17_-_42988004 | 0.21 |
ENST00000586125.1
ENST00000591880.1 |
GFAP
|
glial fibrillary acidic protein |
| chr4_-_71705060 | 0.21 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr2_+_162016916 | 0.21 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr17_+_52978185 | 0.20 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr10_+_14880364 | 0.20 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr2_+_234621551 | 0.20 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr17_+_9745786 | 0.20 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr11_+_86748957 | 0.20 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chr13_+_97928395 | 0.19 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr11_-_64512273 | 0.19 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr10_-_14880002 | 0.19 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr17_-_79359154 | 0.19 |
ENST00000572077.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr14_+_75761099 | 0.19 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr3_-_10028366 | 0.18 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr12_+_50794891 | 0.18 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr6_-_76072719 | 0.17 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr11_-_64511789 | 0.17 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr12_+_7169887 | 0.17 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr12_-_8088773 | 0.16 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr3_-_197024965 | 0.16 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_179322436 | 0.16 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr4_+_81118647 | 0.16 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chrY_-_15591485 | 0.15 |
ENST00000382896.4
ENST00000537580.1 ENST00000540140.1 ENST00000545955.1 ENST00000538878.1 |
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr2_+_234627424 | 0.15 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr2_+_103378472 | 0.15 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr3_+_179322573 | 0.15 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr12_+_51818555 | 0.15 |
ENST00000453097.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr6_+_136172820 | 0.15 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr17_-_49021974 | 0.15 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr1_+_155583012 | 0.15 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr12_-_95397442 | 0.15 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr1_+_45140360 | 0.15 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr22_+_37959647 | 0.15 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr14_-_104387888 | 0.15 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr12_+_50794947 | 0.14 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr5_+_66124590 | 0.14 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr16_-_66952742 | 0.14 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr1_-_17380630 | 0.14 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr11_+_34938119 | 0.14 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr1_+_201924619 | 0.14 |
ENST00000367287.4
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr2_+_220299547 | 0.14 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr11_-_64511575 | 0.13 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr7_+_112090483 | 0.13 |
ENST00000403825.3
ENST00000429071.1 |
IFRD1
|
interferon-related developmental regulator 1 |
| chrX_-_21776281 | 0.13 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr2_+_198365095 | 0.13 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr3_-_113465065 | 0.13 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chrX_-_109683446 | 0.13 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chrX_+_135230712 | 0.13 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr20_+_44462749 | 0.12 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr7_-_91875358 | 0.12 |
ENST00000458177.1
ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr11_-_47470703 | 0.12 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr1_-_157108266 | 0.12 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr15_-_55562451 | 0.12 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr20_+_57875457 | 0.12 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr3_-_179322416 | 0.12 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr1_-_205719295 | 0.12 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr4_+_26321284 | 0.12 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_-_25194963 | 0.11 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr14_-_92572894 | 0.11 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr14_-_21492113 | 0.11 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr4_-_140216948 | 0.11 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_+_198365122 | 0.11 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr10_+_14880157 | 0.11 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr12_-_56753858 | 0.11 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr3_-_151034734 | 0.11 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr7_+_128399002 | 0.11 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr11_-_9336234 | 0.11 |
ENST00000528080.1
|
TMEM41B
|
transmembrane protein 41B |
| chr14_-_104387831 | 0.11 |
ENST00000557040.1
ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2
|
chromosome 14 open reading frame 2 |
| chr20_-_44485835 | 0.11 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr7_+_117864708 | 0.11 |
ENST00000357099.4
ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr2_-_85895295 | 0.11 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr20_-_62130474 | 0.10 |
ENST00000217182.3
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr5_-_131347583 | 0.10 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr6_-_24666819 | 0.10 |
ENST00000341060.3
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr14_-_55369525 | 0.10 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr3_-_197025447 | 0.10 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr7_-_91875109 | 0.10 |
ENST00000412043.2
ENST00000430102.1 ENST00000425073.1 ENST00000394503.2 ENST00000454017.1 ENST00000440209.1 ENST00000413688.1 ENST00000452773.1 ENST00000433016.1 ENST00000394505.2 ENST00000422347.1 ENST00000458493.1 ENST00000425919.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr2_+_210288760 | 0.10 |
ENST00000199940.6
|
MAP2
|
microtubule-associated protein 2 |
| chr9_-_96215822 | 0.10 |
ENST00000375412.5
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr12_+_51818749 | 0.10 |
ENST00000514353.3
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr7_-_72936531 | 0.10 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr10_-_33625154 | 0.10 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr3_-_52860850 | 0.09 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr17_+_19281787 | 0.09 |
ENST00000482850.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr9_-_88874519 | 0.09 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr5_-_131347501 | 0.09 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr2_-_207024134 | 0.09 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_169075554 | 0.09 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr8_-_100905850 | 0.09 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr4_-_65275162 | 0.09 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr7_-_27169801 | 0.09 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr6_+_149068464 | 0.09 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr2_-_198364581 | 0.09 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr5_-_137878887 | 0.09 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr16_-_4292071 | 0.09 |
ENST00000399609.3
|
SRL
|
sarcalumenin |
| chr4_+_48343339 | 0.08 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr12_-_8088871 | 0.08 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr5_-_133340326 | 0.08 |
ENST00000425992.1
ENST00000395044.3 ENST00000395047.2 |
VDAC1
|
voltage-dependent anion channel 1 |
| chr7_-_151433393 | 0.08 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr5_-_149792295 | 0.08 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr11_-_47470682 | 0.08 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr6_-_131299929 | 0.08 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr7_+_91875508 | 0.08 |
ENST00000265742.3
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1 |
| chr16_-_58328884 | 0.08 |
ENST00000569079.1
|
PRSS54
|
protease, serine, 54 |
| chr7_-_151433342 | 0.08 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr2_+_120189422 | 0.08 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr20_+_57875758 | 0.08 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr8_-_100905925 | 0.08 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr7_-_140179276 | 0.08 |
ENST00000443720.2
ENST00000255977.2 |
MKRN1
|
makorin ring finger protein 1 |
| chr20_+_57875658 | 0.08 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr7_-_140624499 | 0.08 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr15_+_43885799 | 0.07 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr19_-_55874611 | 0.07 |
ENST00000424985.3
|
FAM71E2
|
family with sequence similarity 71, member E2 |
| chr20_-_60718430 | 0.07 |
ENST00000370873.4
ENST00000370858.3 |
PSMA7
|
proteasome (prosome, macropain) subunit, alpha type, 7 |
| chr1_-_104239076 | 0.07 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr1_-_104238912 | 0.07 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chrX_+_72223352 | 0.07 |
ENST00000373521.2
ENST00000538388.1 |
PABPC1L2B
|
poly(A) binding protein, cytoplasmic 1-like 2B |
| chr22_+_20104947 | 0.07 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr16_-_49698136 | 0.07 |
ENST00000535559.1
|
ZNF423
|
zinc finger protein 423 |
| chr15_+_43985084 | 0.07 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr4_+_41614909 | 0.07 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr2_+_219745020 | 0.07 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr1_+_26348259 | 0.07 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr11_-_63993690 | 0.07 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr6_-_24667180 | 0.07 |
ENST00000545995.1
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr6_+_107077471 | 0.07 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr1_+_156549519 | 0.07 |
ENST00000368236.3
|
TTC24
|
tetratricopeptide repeat domain 24 |
| chr20_+_13765596 | 0.06 |
ENST00000378106.5
ENST00000463598.1 |
NDUFAF5
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
| chr2_-_287299 | 0.06 |
ENST00000405290.1
|
FAM150B
|
family with sequence similarity 150, member B |
| chr16_-_30134524 | 0.06 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr17_-_36760865 | 0.06 |
ENST00000584266.1
|
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr2_-_207023918 | 0.06 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr6_-_107077347 | 0.06 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr1_+_32084641 | 0.06 |
ENST00000373706.5
|
HCRTR1
|
hypocretin (orexin) receptor 1 |
| chr10_+_22605304 | 0.06 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr1_-_157108130 | 0.06 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr14_-_23791484 | 0.06 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr7_-_127672146 | 0.06 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr17_-_9939854 | 0.06 |
ENST00000584146.2
|
GAS7
|
growth arrest-specific 7 |
| chr5_-_169626104 | 0.06 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr10_-_120938303 | 0.06 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr8_-_70745575 | 0.05 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr3_-_46000146 | 0.05 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr12_-_15865844 | 0.05 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr2_-_25194476 | 0.05 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr19_+_35629702 | 0.05 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr8_-_100905363 | 0.05 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr11_-_9336117 | 0.05 |
ENST00000527813.1
ENST00000533723.1 |
TMEM41B
|
transmembrane protein 41B |
| chr11_-_63993601 | 0.05 |
ENST00000545812.1
ENST00000394547.3 ENST00000317459.6 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr1_-_226129189 | 0.05 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr15_+_78441663 | 0.05 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr19_+_45251804 | 0.05 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr14_-_48144157 | 0.05 |
ENST00000399232.2
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr3_+_107096188 | 0.05 |
ENST00000261058.1
|
CCDC54
|
coiled-coil domain containing 54 |
| chr17_-_9939935 | 0.05 |
ENST00000580043.1
|
GAS7
|
growth arrest-specific 7 |
| chr1_-_226129083 | 0.04 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr1_-_234667504 | 0.04 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr15_+_43985725 | 0.04 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr10_+_51371390 | 0.04 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr10_-_50970382 | 0.04 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr3_+_129247479 | 0.04 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr2_+_85804614 | 0.04 |
ENST00000263864.5
ENST00000409760.1 |
VAMP8
|
vesicle-associated membrane protein 8 |
| chr1_+_16062820 | 0.04 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr17_-_32484313 | 0.04 |
ENST00000359872.6
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr7_+_129007964 | 0.04 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr6_+_107077435 | 0.04 |
ENST00000369046.4
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr3_-_49459878 | 0.04 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0045362 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.5 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 0.7 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.4 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.6 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.1 | 0.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.3 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.4 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0072554 | blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:1903276 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.5 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.3 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.4 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.7 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.2 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |