Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
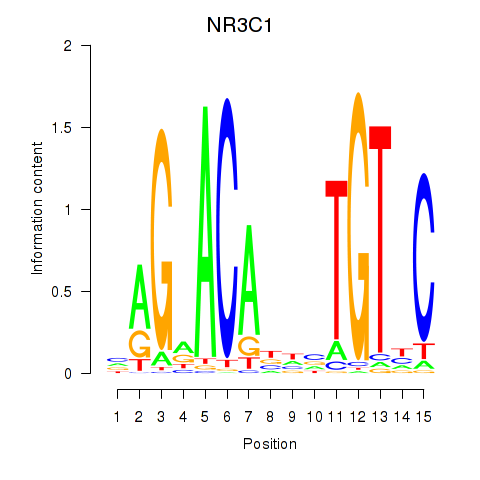
Results for NR3C1
Z-value: 0.99
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.10 | nuclear receptor subfamily 3 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg19_v2_chr5_-_142782862_142782894 | 0.83 | 4.1e-02 | Click! |
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_47352477 | 2.78 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr1_-_27998689 | 2.06 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr8_-_145060593 | 1.13 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr2_-_191878162 | 1.05 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr12_-_7245125 | 0.72 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr7_-_95225768 | 0.69 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr19_+_15783879 | 0.68 |
ENST00000551607.1
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12 |
| chr2_+_114163945 | 0.63 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr4_+_9446156 | 0.62 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr1_+_218683438 | 0.62 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr20_+_52824367 | 0.57 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr14_+_24641062 | 0.57 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr12_-_7244469 | 0.56 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr11_+_57365150 | 0.52 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr12_-_7245152 | 0.48 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr3_+_169490606 | 0.47 |
ENST00000349841.5
|
MYNN
|
myoneurin |
| chr6_+_26045603 | 0.46 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr8_-_90993869 | 0.46 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr13_-_26452679 | 0.45 |
ENST00000596729.1
|
AL138815.2
|
Uncharacterized protein |
| chr12_-_8693469 | 0.44 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr6_+_77484663 | 0.44 |
ENST00000607287.1
|
RP11-354K4.2
|
RP11-354K4.2 |
| chr2_-_113594279 | 0.43 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr8_+_91013577 | 0.42 |
ENST00000220764.2
|
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr3_-_75834722 | 0.42 |
ENST00000471541.2
|
ZNF717
|
zinc finger protein 717 |
| chr13_-_108867846 | 0.41 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr19_+_1495362 | 0.41 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr2_-_191399073 | 0.41 |
ENST00000421038.1
|
TMEM194B
|
transmembrane protein 194B |
| chr9_-_15307200 | 0.40 |
ENST00000506891.1
ENST00000541445.1 ENST00000512701.2 ENST00000380850.4 ENST00000297615.5 ENST00000355694.2 |
TTC39B
|
tetratricopeptide repeat domain 39B |
| chr5_+_158690089 | 0.40 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr19_+_4229495 | 0.39 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr11_+_18287721 | 0.39 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr19_-_30199516 | 0.38 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr5_+_82373379 | 0.38 |
ENST00000396027.4
ENST00000511817.1 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr12_-_7245018 | 0.37 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr5_+_82373317 | 0.36 |
ENST00000282268.3
ENST00000338635.6 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr5_+_43033818 | 0.36 |
ENST00000607830.1
|
CTD-2035E11.4
|
CTD-2035E11.4 |
| chr9_-_37384431 | 0.36 |
ENST00000452923.1
|
RP11-397D12.4
|
RP11-397D12.4 |
| chr12_-_8693539 | 0.35 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr11_+_114271367 | 0.35 |
ENST00000544582.1
ENST00000545678.1 |
RBM7
|
RNA binding motif protein 7 |
| chr5_-_55777586 | 0.35 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr7_+_107220660 | 0.35 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr12_-_7245080 | 0.35 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr7_-_139727118 | 0.34 |
ENST00000484111.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr4_+_75310851 | 0.34 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr14_-_92333873 | 0.34 |
ENST00000435962.2
|
TC2N
|
tandem C2 domains, nuclear |
| chr17_+_70036164 | 0.34 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr8_+_91013676 | 0.33 |
ENST00000519410.1
ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr1_-_235098861 | 0.33 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr20_-_62493217 | 0.33 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr11_+_105948216 | 0.33 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr12_-_110486281 | 0.33 |
ENST00000546627.1
|
C12orf76
|
chromosome 12 open reading frame 76 |
| chr12_+_104697504 | 0.31 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr9_-_34372830 | 0.31 |
ENST00000379142.3
|
KIAA1161
|
KIAA1161 |
| chr19_+_46002868 | 0.31 |
ENST00000396735.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr2_+_170440844 | 0.31 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr16_-_29275800 | 0.30 |
ENST00000567984.1
|
RP11-426C22.6
|
RP11-426C22.6 |
| chr12_-_105352080 | 0.30 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr8_-_102987548 | 0.30 |
ENST00000520690.1
|
NCALD
|
neurocalcin delta |
| chr18_+_61442629 | 0.29 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr4_+_75311019 | 0.29 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr15_+_49913201 | 0.29 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr12_-_123459105 | 0.29 |
ENST00000543935.1
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr11_-_77734260 | 0.29 |
ENST00000353172.5
|
KCTD14
|
potassium channel tetramerization domain containing 14 |
| chr2_-_61389240 | 0.28 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr4_+_75480629 | 0.28 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr4_+_90033968 | 0.28 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr10_-_18948208 | 0.28 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr10_+_120863587 | 0.28 |
ENST00000535029.1
ENST00000361432.2 ENST00000544016.1 |
FAM45A
|
family with sequence similarity 45, member A |
| chr1_-_74663825 | 0.28 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr15_+_65204075 | 0.27 |
ENST00000380230.3
ENST00000357698.3 ENST00000395720.1 ENST00000496660.1 ENST00000319580.8 |
ANKDD1A
|
ankyrin repeat and death domain containing 1A |
| chr10_+_27793257 | 0.27 |
ENST00000375802.3
|
RAB18
|
RAB18, member RAS oncogene family |
| chr8_+_38614754 | 0.27 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr11_-_790060 | 0.27 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr14_-_24616426 | 0.27 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr17_+_57233087 | 0.27 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr6_-_27835357 | 0.26 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr2_-_200715573 | 0.26 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr12_+_9144626 | 0.26 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr2_-_17981462 | 0.26 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chrX_-_154255143 | 0.26 |
ENST00000453950.1
ENST00000423959.1 |
F8
|
coagulation factor VIII, procoagulant component |
| chr6_+_76330355 | 0.26 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr10_+_81466084 | 0.26 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr2_+_208104497 | 0.26 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr6_-_106773291 | 0.25 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr7_-_130353553 | 0.25 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr2_-_170430277 | 0.25 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr1_+_70876926 | 0.25 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr3_+_169491171 | 0.25 |
ENST00000356716.4
|
MYNN
|
myoneurin |
| chr5_-_86708670 | 0.25 |
ENST00000504878.1
|
CCNH
|
cyclin H |
| chr16_+_56716336 | 0.25 |
ENST00000394485.4
ENST00000562939.1 |
MT1X
|
metallothionein 1X |
| chr15_+_33010175 | 0.25 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr14_+_24641820 | 0.25 |
ENST00000560501.1
|
REC8
|
REC8 meiotic recombination protein |
| chr20_+_12989596 | 0.24 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr1_+_6511651 | 0.24 |
ENST00000434576.1
|
ESPN
|
espin |
| chr15_-_56757329 | 0.24 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr16_+_75256507 | 0.24 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr16_+_1578674 | 0.24 |
ENST00000253934.5
|
TMEM204
|
transmembrane protein 204 |
| chr8_+_1919481 | 0.24 |
ENST00000518539.2
|
KBTBD11-OT1
|
KBTBD11 overlapping transcript 1 (non-protein coding) |
| chr14_+_24616588 | 0.24 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr22_+_50981079 | 0.24 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr1_+_155583012 | 0.24 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr19_+_56905024 | 0.24 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr15_+_55700741 | 0.24 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr19_-_2282167 | 0.24 |
ENST00000342063.3
|
C19orf35
|
chromosome 19 open reading frame 35 |
| chr12_-_88535842 | 0.24 |
ENST00000550962.1
ENST00000552810.1 |
CEP290
|
centrosomal protein 290kDa |
| chr8_-_22014339 | 0.24 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr4_+_15779901 | 0.23 |
ENST00000226279.3
|
CD38
|
CD38 molecule |
| chr10_+_99627889 | 0.23 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr9_-_88356789 | 0.23 |
ENST00000357081.3
ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr2_+_170440902 | 0.23 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr6_-_133035185 | 0.23 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr2_+_234637754 | 0.23 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_-_169769787 | 0.23 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr3_-_138312971 | 0.23 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr5_+_162887556 | 0.23 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr3_+_169490834 | 0.23 |
ENST00000392733.1
|
MYNN
|
myoneurin |
| chr11_-_102323489 | 0.23 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr17_-_46178527 | 0.23 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr1_+_62901968 | 0.22 |
ENST00000452143.1
ENST00000442679.1 ENST00000371146.1 |
USP1
|
ubiquitin specific peptidase 1 |
| chr3_+_142720366 | 0.22 |
ENST00000493782.1
ENST00000397933.2 ENST00000473835.2 ENST00000493598.2 |
U2SURP
|
U2 snRNP-associated SURP domain containing |
| chr19_+_21324863 | 0.22 |
ENST00000598331.1
|
ZNF431
|
zinc finger protein 431 |
| chr8_-_121457332 | 0.22 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr5_+_94982435 | 0.22 |
ENST00000511684.1
ENST00000380005.4 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr8_-_82754193 | 0.22 |
ENST00000519817.1
ENST00000521773.1 ENST00000523757.1 |
SNX16
|
sorting nexin 16 |
| chr8_-_22014255 | 0.22 |
ENST00000424267.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr11_+_110300607 | 0.21 |
ENST00000260270.2
|
FDX1
|
ferredoxin 1 |
| chr13_-_96329048 | 0.21 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr1_+_45965725 | 0.21 |
ENST00000401061.4
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr7_-_108097144 | 0.21 |
ENST00000418239.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr9_+_126777676 | 0.21 |
ENST00000488674.2
|
LHX2
|
LIM homeobox 2 |
| chr14_+_57735725 | 0.21 |
ENST00000431972.2
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr10_-_111713633 | 0.21 |
ENST00000538668.1
ENST00000369657.1 ENST00000369655.1 |
RP11-451M19.3
|
Uncharacterized protein |
| chr14_-_45603657 | 0.21 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr1_-_109203997 | 0.21 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr16_+_84209738 | 0.20 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr4_-_147443043 | 0.20 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr16_+_226658 | 0.20 |
ENST00000320868.5
ENST00000397797.1 |
HBA1
|
hemoglobin, alpha 1 |
| chr5_-_39219641 | 0.20 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr7_-_93519471 | 0.20 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr19_-_37958086 | 0.20 |
ENST00000592490.1
ENST00000392149.2 |
ZNF569
|
zinc finger protein 569 |
| chr3_-_42846021 | 0.20 |
ENST00000321331.7
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr1_+_149230680 | 0.20 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chr17_+_40950797 | 0.20 |
ENST00000588408.1
ENST00000585355.1 |
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr8_-_82754427 | 0.20 |
ENST00000353788.4
ENST00000520618.1 ENST00000518183.1 ENST00000396330.2 ENST00000519119.1 ENST00000345957.4 |
SNX16
|
sorting nexin 16 |
| chr2_-_75788428 | 0.20 |
ENST00000432649.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr16_+_89238149 | 0.20 |
ENST00000289746.2
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr1_-_222885770 | 0.20 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr16_-_84150410 | 0.20 |
ENST00000569907.1
|
MBTPS1
|
membrane-bound transcription factor peptidase, site 1 |
| chr8_-_103136481 | 0.20 |
ENST00000524209.1
ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD
|
neurocalcin delta |
| chr8_+_110551925 | 0.20 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr5_-_39219705 | 0.19 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr3_-_42845922 | 0.19 |
ENST00000452906.2
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr11_-_105948040 | 0.19 |
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr2_+_26785409 | 0.19 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr7_+_6655225 | 0.19 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr3_-_138313161 | 0.19 |
ENST00000489254.1
ENST00000474781.1 ENST00000462419.1 ENST00000264982.3 |
CEP70
|
centrosomal protein 70kDa |
| chr19_+_53868940 | 0.19 |
ENST00000474037.1
ENST00000491101.1 ENST00000467003.1 ENST00000475179.1 ENST00000593918.1 |
ZNF525
|
zinc finger protein 525 |
| chr7_-_95064264 | 0.19 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr19_-_37019562 | 0.19 |
ENST00000523638.1
|
ZNF260
|
zinc finger protein 260 |
| chr17_-_18585131 | 0.19 |
ENST00000443457.1
ENST00000583002.1 |
ZNF286B
|
zinc finger protein 286B |
| chr14_-_38036271 | 0.19 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr16_-_3285049 | 0.19 |
ENST00000575948.1
|
ZNF200
|
zinc finger protein 200 |
| chrX_-_80377118 | 0.19 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr3_+_122044084 | 0.19 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr10_+_60145155 | 0.18 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr21_+_35553045 | 0.18 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr11_+_34663913 | 0.18 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr18_+_47087390 | 0.18 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr2_+_10861775 | 0.18 |
ENST00000272238.4
ENST00000381661.3 |
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 |
| chr9_-_19065082 | 0.18 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr3_+_52448539 | 0.18 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr5_+_94982558 | 0.18 |
ENST00000311364.4
ENST00000458310.1 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr11_+_18287801 | 0.18 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chrX_+_71401570 | 0.18 |
ENST00000496835.2
ENST00000446576.1 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr1_+_90460661 | 0.18 |
ENST00000340281.4
ENST00000361911.5 ENST00000370447.3 ENST00000455342.2 |
ZNF326
|
zinc finger protein 326 |
| chr3_-_112738490 | 0.18 |
ENST00000393857.2
|
C3orf17
|
chromosome 3 open reading frame 17 |
| chr15_-_34447023 | 0.18 |
ENST00000560310.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr1_-_241799232 | 0.18 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr3_+_188817891 | 0.18 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr5_+_125935960 | 0.18 |
ENST00000297540.4
|
PHAX
|
phosphorylated adaptor for RNA export |
| chr19_-_54579813 | 0.18 |
ENST00000446034.2
|
TARM1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr14_+_96671016 | 0.18 |
ENST00000542454.2
ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2
RP11-404P21.8
|
bradykinin receptor B2 Uncharacterized protein |
| chr5_-_86708833 | 0.18 |
ENST00000256897.4
|
CCNH
|
cyclin H |
| chr4_+_55095428 | 0.18 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr11_-_627143 | 0.18 |
ENST00000176195.3
|
SCT
|
secretin |
| chr14_+_88851874 | 0.17 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chr10_+_25305524 | 0.17 |
ENST00000524413.1
ENST00000376356.4 |
THNSL1
|
threonine synthase-like 1 (S. cerevisiae) |
| chr19_-_53662257 | 0.17 |
ENST00000599096.1
ENST00000334197.7 ENST00000597183.1 ENST00000601804.1 ENST00000601469.2 ENST00000452676.2 |
ZNF347
|
zinc finger protein 347 |
| chr3_+_9944303 | 0.17 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr15_-_55700216 | 0.17 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chrX_-_71458802 | 0.17 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr2_+_242681835 | 0.17 |
ENST00000437164.1
ENST00000454048.1 ENST00000417686.1 |
D2HGDH
|
D-2-hydroxyglutarate dehydrogenase |
| chr13_-_36920872 | 0.17 |
ENST00000451493.1
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr1_-_27816556 | 0.17 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr8_-_82644562 | 0.17 |
ENST00000520604.1
ENST00000521742.1 ENST00000520635.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr6_+_127898312 | 0.17 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr19_+_57922521 | 0.17 |
ENST00000307658.7
ENST00000601808.1 |
ZNF17
|
zinc finger protein 17 |
| chr14_+_88852059 | 0.17 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr17_-_57232525 | 0.17 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr6_+_31638156 | 0.17 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 1.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 2.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.2 | 0.5 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.4 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.5 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.5 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.1 | 0.3 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.6 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.2 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.2 | GO:0090274 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 1.0 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.3 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.3 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.7 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.3 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.3 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.3 | GO:0036018 | response to erythropoietin(GO:0036017) cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:1901558 | regulation of cholesterol transporter activity(GO:0060694) response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.0 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.1 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 1.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.7 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.3 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.0 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.2 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.1 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0070417 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0035811 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:2000523 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:1902904 | late endosomal microautophagy(GO:0061738) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.8 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.2 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.1 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0070702 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.6 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.0 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.2 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 0.5 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.2 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.1 | 0.2 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.0 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 1.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0034617 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005501 | retinoid binding(GO:0005501) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |