Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
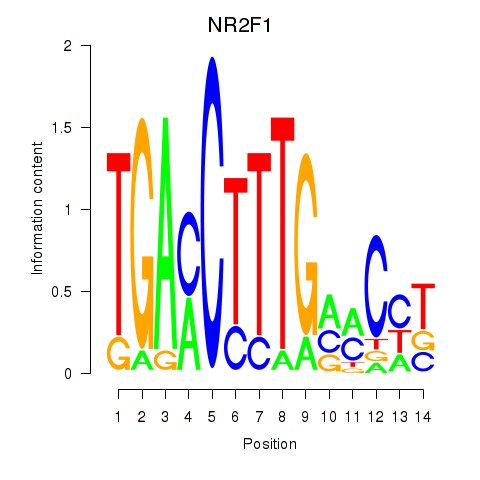
Results for NR2F1
Z-value: 0.70
Transcription factors associated with NR2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F1
|
ENSG00000175745.7 | nuclear receptor subfamily 2 group F member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F1 | hg19_v2_chr5_+_92919043_92919082 | 0.72 | 1.1e-01 | Click! |
Activity profile of NR2F1 motif
Sorted Z-values of NR2F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48867291 | 0.59 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr2_+_220306238 | 0.52 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr8_-_37594944 | 0.50 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr1_+_23695680 | 0.48 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr9_+_6716478 | 0.48 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr6_+_31926857 | 0.46 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr16_+_81272287 | 0.44 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr11_-_45939374 | 0.42 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr6_-_33771757 | 0.42 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr19_+_39616410 | 0.42 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr19_-_39303576 | 0.41 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr11_-_45939565 | 0.39 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr3_+_11267691 | 0.36 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr16_-_88752889 | 0.36 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr12_-_31158902 | 0.35 |
ENST00000544329.1
ENST00000418254.2 ENST00000222396.5 |
RP11-551L14.4
|
RP11-551L14.4 |
| chr19_+_6464502 | 0.35 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr11_+_63974135 | 0.35 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr16_-_69448 | 0.33 |
ENST00000326592.9
|
WASH4P
|
WAS protein family homolog 4 pseudogene |
| chr2_+_27282134 | 0.33 |
ENST00000441931.1
|
AGBL5
|
ATP/GTP binding protein-like 5 |
| chr17_-_7531121 | 0.33 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr16_+_30710462 | 0.31 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr5_-_180235755 | 0.31 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr7_-_642261 | 0.30 |
ENST00000400758.2
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr19_-_1174226 | 0.29 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr12_-_56367101 | 0.28 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr19_-_48867171 | 0.28 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr12_+_49717081 | 0.27 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr17_+_77893135 | 0.27 |
ENST00000574526.1
ENST00000572353.1 |
RP11-353N14.4
|
RP11-353N14.4 |
| chr17_-_74489215 | 0.27 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr7_+_99036996 | 0.26 |
ENST00000441580.1
ENST00000412686.1 |
CPSF4
|
cleavage and polyadenylation specific factor 4, 30kDa |
| chr19_+_12780512 | 0.26 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr7_-_37956409 | 0.26 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr19_+_58790314 | 0.26 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr19_-_36304201 | 0.25 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr18_+_54318566 | 0.25 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr14_+_24590560 | 0.25 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr11_-_26743546 | 0.25 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr16_+_2255841 | 0.25 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr16_+_83932684 | 0.24 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr2_+_219433281 | 0.24 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr6_+_143999185 | 0.24 |
ENST00000542769.1
ENST00000397980.3 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr17_-_47723943 | 0.24 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr7_+_143080063 | 0.24 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr19_+_6464243 | 0.23 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr19_+_10222189 | 0.23 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chr2_+_73441350 | 0.23 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr9_+_129097520 | 0.22 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr3_+_9691117 | 0.22 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr22_-_29784519 | 0.22 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr1_-_23520755 | 0.22 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr9_+_129097479 | 0.22 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr1_-_183538319 | 0.22 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr1_+_62439037 | 0.22 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr22_+_27068704 | 0.21 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr11_+_67798090 | 0.21 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr1_-_55089191 | 0.21 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr5_+_1801503 | 0.21 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr6_+_44094627 | 0.21 |
ENST00000259746.9
|
TMEM63B
|
transmembrane protein 63B |
| chr19_-_14201776 | 0.21 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr7_-_74489609 | 0.21 |
ENST00000329959.4
ENST00000503250.2 ENST00000543840.1 |
WBSCR16
|
Williams-Beuren syndrome chromosome region 16 |
| chr20_+_34802295 | 0.21 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr2_-_70780572 | 0.20 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr9_-_16705069 | 0.20 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr12_+_49717019 | 0.20 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr11_-_72493574 | 0.20 |
ENST00000536290.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr22_+_24105394 | 0.20 |
ENST00000305199.5
ENST00000382821.3 |
C22orf15
|
chromosome 22 open reading frame 15 |
| chr8_-_135652051 | 0.20 |
ENST00000522257.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr19_+_11546093 | 0.19 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr1_+_44679159 | 0.19 |
ENST00000315913.5
ENST00000372289.2 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr16_+_2255710 | 0.19 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr1_+_43855560 | 0.19 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr19_+_10362882 | 0.19 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr1_+_110162448 | 0.19 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr4_-_40477766 | 0.19 |
ENST00000507180.1
|
RBM47
|
RNA binding motif protein 47 |
| chr8_+_144816303 | 0.19 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr12_-_48214345 | 0.19 |
ENST00000433685.1
ENST00000447463.1 ENST00000427332.2 |
HDAC7
|
histone deacetylase 7 |
| chr11_+_67798114 | 0.18 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr17_-_16472483 | 0.18 |
ENST00000395824.1
ENST00000448349.2 ENST00000395825.3 |
ZNF287
|
zinc finger protein 287 |
| chr12_-_6665200 | 0.18 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr10_-_104211294 | 0.18 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr6_-_31926629 | 0.18 |
ENST00000375425.5
ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE
|
negative elongation factor complex member E |
| chr3_-_194393206 | 0.18 |
ENST00000265245.5
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr19_-_9968816 | 0.18 |
ENST00000590841.1
|
OLFM2
|
olfactomedin 2 |
| chr2_-_85581623 | 0.18 |
ENST00000449375.1
ENST00000409984.2 ENST00000457495.2 ENST00000263854.6 |
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr1_-_29508499 | 0.18 |
ENST00000373795.4
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr7_-_35293740 | 0.18 |
ENST00000408931.3
|
TBX20
|
T-box 20 |
| chr2_+_220363579 | 0.18 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr1_-_91328477 | 0.18 |
ENST00000606898.1
|
RP4-665J23.4
|
RP4-665J23.4 |
| chr11_-_1776176 | 0.17 |
ENST00000429746.1
|
CTSD
|
cathepsin D |
| chr8_-_16859690 | 0.17 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr1_+_181003067 | 0.17 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr8_+_22853345 | 0.17 |
ENST00000522948.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr11_+_2920951 | 0.17 |
ENST00000347936.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr16_+_21244986 | 0.17 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr17_+_73606766 | 0.16 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chr4_-_109541539 | 0.16 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr19_+_11546153 | 0.16 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr19_-_39390440 | 0.16 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr7_+_192969 | 0.16 |
ENST00000313766.5
|
FAM20C
|
family with sequence similarity 20, member C |
| chr11_+_118868830 | 0.16 |
ENST00000334418.1
|
CCDC84
|
coiled-coil domain containing 84 |
| chr1_-_229569834 | 0.16 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr17_-_4852332 | 0.16 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr22_+_47158518 | 0.16 |
ENST00000337137.4
ENST00000380995.1 ENST00000407381.3 |
TBC1D22A
|
TBC1 domain family, member 22A |
| chr17_-_4852243 | 0.16 |
ENST00000225655.5
|
PFN1
|
profilin 1 |
| chr13_+_76413852 | 0.16 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr19_+_11546440 | 0.16 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr19_-_6767431 | 0.16 |
ENST00000437152.3
ENST00000597687.1 |
SH2D3A
|
SH2 domain containing 3A |
| chr9_+_127213423 | 0.16 |
ENST00000334810.1
|
GPR144
|
G protein-coupled receptor 144 |
| chr8_-_8639956 | 0.16 |
ENST00000522213.1
|
RP11-211C9.1
|
RP11-211C9.1 |
| chr12_-_109219937 | 0.16 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr1_-_29508321 | 0.16 |
ENST00000546138.1
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr2_+_10442993 | 0.15 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr4_-_141075330 | 0.15 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr1_+_44679113 | 0.15 |
ENST00000361745.6
ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr6_+_43737939 | 0.15 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr22_+_19950060 | 0.15 |
ENST00000449653.1
|
COMT
|
catechol-O-methyltransferase |
| chr7_+_150782945 | 0.15 |
ENST00000463381.1
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr11_-_8816375 | 0.15 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr19_-_633576 | 0.15 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr12_+_113659234 | 0.15 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr12_+_121416489 | 0.15 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr8_-_144886321 | 0.15 |
ENST00000526832.1
|
SCRIB
|
scribbled planar cell polarity protein |
| chr2_-_3584430 | 0.15 |
ENST00000438482.1
ENST00000422961.1 |
AC108488.4
|
AC108488.4 |
| chr11_+_63137251 | 0.15 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr9_-_39239171 | 0.15 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr7_+_75911902 | 0.15 |
ENST00000413003.1
|
SRRM3
|
serine/arginine repetitive matrix 3 |
| chr12_+_121416340 | 0.15 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr19_-_1021113 | 0.15 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr6_+_7108210 | 0.15 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr3_+_184055240 | 0.14 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr16_+_2076869 | 0.14 |
ENST00000424542.2
ENST00000432365.2 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr11_+_72929319 | 0.14 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr1_+_196857144 | 0.14 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr12_-_6960407 | 0.14 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr16_+_4475806 | 0.14 |
ENST00000262375.6
ENST00000355296.4 ENST00000431375.2 ENST00000574895.1 |
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr6_+_54173227 | 0.14 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr4_+_159593271 | 0.14 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr12_-_49245936 | 0.14 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr1_-_151299842 | 0.14 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr7_+_97840739 | 0.14 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr19_-_42758040 | 0.14 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr9_+_104296243 | 0.14 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr6_+_30131318 | 0.14 |
ENST00000376688.1
|
TRIM15
|
tripartite motif containing 15 |
| chr20_-_62601218 | 0.13 |
ENST00000369888.1
|
ZNF512B
|
zinc finger protein 512B |
| chr9_+_104296163 | 0.13 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr22_+_47158578 | 0.13 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr12_+_58013693 | 0.13 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr17_-_36105009 | 0.13 |
ENST00000560016.1
ENST00000427275.2 ENST00000561193.1 |
HNF1B
|
HNF1 homeobox B |
| chr7_-_65447192 | 0.13 |
ENST00000421103.1
ENST00000345660.6 ENST00000304895.4 |
GUSB
|
glucuronidase, beta |
| chr22_+_25348671 | 0.13 |
ENST00000406486.4
|
KIAA1671
|
KIAA1671 |
| chr12_+_132413798 | 0.13 |
ENST00000440818.2
ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1
|
pseudouridylate synthase 1 |
| chr22_+_25003626 | 0.13 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_-_85581701 | 0.13 |
ENST00000295802.4
|
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr2_-_73460334 | 0.13 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr11_+_7595136 | 0.13 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr15_+_101389945 | 0.13 |
ENST00000561231.1
ENST00000559331.1 ENST00000558254.1 |
RP11-66B24.2
|
RP11-66B24.2 |
| chr11_+_46366799 | 0.12 |
ENST00000532868.2
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr2_-_70780770 | 0.12 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr5_-_42812143 | 0.12 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr15_+_91449971 | 0.12 |
ENST00000557865.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr16_+_29840929 | 0.12 |
ENST00000566252.1
|
MVP
|
major vault protein |
| chr15_-_75660919 | 0.12 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr14_+_101295948 | 0.12 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_+_39390587 | 0.12 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr17_+_75446819 | 0.12 |
ENST00000541152.2
ENST00000591704.1 |
SEPT9
|
septin 9 |
| chr7_-_100844193 | 0.12 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr1_-_36235559 | 0.12 |
ENST00000251195.5
|
CLSPN
|
claspin |
| chr6_+_37897735 | 0.12 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr16_+_67207872 | 0.12 |
ENST00000563258.1
ENST00000568146.1 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr6_+_7107999 | 0.12 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr10_-_74114714 | 0.12 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr16_-_1494490 | 0.11 |
ENST00000389176.3
ENST00000409671.1 |
CCDC154
|
coiled-coil domain containing 154 |
| chr1_-_53793725 | 0.11 |
ENST00000371454.2
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr17_+_75446629 | 0.11 |
ENST00000588958.2
ENST00000586128.1 |
SEPT9
|
septin 9 |
| chr3_-_49837254 | 0.11 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr22_+_25003606 | 0.11 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr16_-_30997533 | 0.11 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr11_+_64073699 | 0.11 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr12_+_57854274 | 0.11 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr17_-_8198636 | 0.11 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr10_+_123922941 | 0.11 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr17_+_27071002 | 0.11 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr11_-_71750865 | 0.11 |
ENST00000544129.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr16_+_2880254 | 0.11 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr5_+_140593509 | 0.11 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr16_+_67207838 | 0.11 |
ENST00000566871.1
ENST00000268605.7 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr11_+_72929402 | 0.11 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr19_-_8408139 | 0.10 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr3_+_10290596 | 0.10 |
ENST00000448281.2
|
TATDN2
|
TatD DNase domain containing 2 |
| chr3_+_128444994 | 0.10 |
ENST00000482525.1
|
RAB7A
|
RAB7A, member RAS oncogene family |
| chr1_-_45253377 | 0.10 |
ENST00000372207.3
|
BEST4
|
bestrophin 4 |
| chr3_-_50336278 | 0.10 |
ENST00000359051.3
ENST00000417393.1 ENST00000442620.1 ENST00000452674.1 |
HYAL3
NAT6
|
hyaluronoglucosaminidase 3 N-acetyltransferase 6 (GCN5-related) |
| chr1_-_6420737 | 0.10 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr8_-_144815966 | 0.10 |
ENST00000388913.3
|
FAM83H
|
family with sequence similarity 83, member H |
| chrX_+_33744503 | 0.10 |
ENST00000439992.1
|
RP11-305F18.1
|
RP11-305F18.1 |
| chr2_+_219135115 | 0.10 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr19_-_38806560 | 0.10 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr20_+_43160458 | 0.10 |
ENST00000372889.1
ENST00000372887.1 ENST00000372882.3 |
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr16_+_3019552 | 0.10 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr1_-_23521222 | 0.10 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.5 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.3 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.2 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.1 | 0.2 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.3 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.1 | 0.1 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.4 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.2 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.1 | 0.2 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.2 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.0 | 0.2 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.3 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.4 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.5 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.4 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.2 | GO:0050666 | cellular response to phosphate starvation(GO:0016036) regulation of sulfur amino acid metabolic process(GO:0031335) positive regulation of sulfur amino acid metabolic process(GO:0031337) regulation of homocysteine metabolic process(GO:0050666) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0044416 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.1 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.0 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.0 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.3 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |