Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
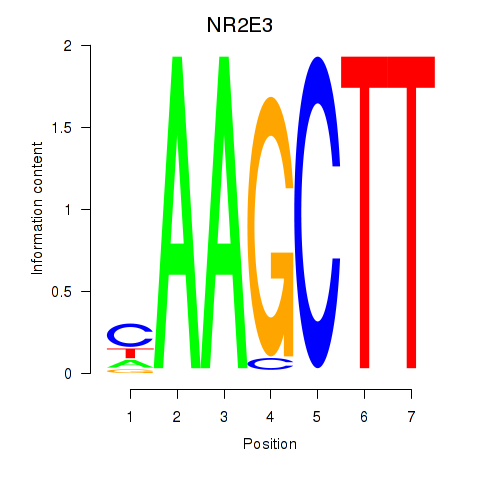
Results for NR2E3
Z-value: 1.06
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000031544.10 | nuclear receptor subfamily 2 group E member 3 |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_66317967 | 0.73 |
ENST00000601398.1
|
AC090673.2
|
Uncharacterized protein |
| chr18_-_33047039 | 0.67 |
ENST00000591141.1
ENST00000586741.1 |
RP11-322E11.5
|
RP11-322E11.5 |
| chr3_-_186262166 | 0.59 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr13_+_51913819 | 0.54 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr12_-_111142729 | 0.52 |
ENST00000546713.1
|
HVCN1
|
hydrogen voltage-gated channel 1 |
| chr3_+_69915363 | 0.48 |
ENST00000451708.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr3_-_195310802 | 0.45 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr11_+_115498761 | 0.44 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr3_+_141105235 | 0.42 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr16_+_67918708 | 0.41 |
ENST00000339176.3
ENST00000576758.1 |
NRN1L
|
neuritin 1-like |
| chr16_-_75569068 | 0.40 |
ENST00000336257.3
ENST00000565039.1 |
CHST5
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 |
| chr3_-_185826718 | 0.39 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chrX_-_27417088 | 0.37 |
ENST00000608735.1
ENST00000422048.1 |
RP11-268G12.1
|
RP11-268G12.1 |
| chr14_+_54976603 | 0.36 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr6_+_148593425 | 0.35 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr20_+_58533471 | 0.34 |
ENST00000244047.5
ENST00000348616.4 |
CDH26
|
cadherin 26 |
| chr10_-_45474237 | 0.34 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chrY_+_15418467 | 0.33 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr5_+_138210919 | 0.33 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr11_-_71639446 | 0.33 |
ENST00000534704.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr2_-_132589601 | 0.31 |
ENST00000437330.1
|
AC103564.7
|
AC103564.7 |
| chr15_+_75640068 | 0.31 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr7_-_17500294 | 0.31 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr3_+_69915385 | 0.30 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_+_145780662 | 0.30 |
ENST00000423031.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr8_-_95449155 | 0.30 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr4_+_71588372 | 0.30 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_+_110749143 | 0.29 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr8_+_77318769 | 0.29 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr10_-_101825151 | 0.29 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr13_+_76334498 | 0.29 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr1_-_179834311 | 0.29 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr12_-_99548524 | 0.29 |
ENST00000549558.2
ENST00000550693.2 ENST00000549493.2 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr15_+_42696992 | 0.27 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chr1_+_168250194 | 0.27 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr2_-_207082748 | 0.26 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr1_+_200638629 | 0.26 |
ENST00000568695.1
|
RP11-92G12.3
|
RP11-92G12.3 |
| chr2_-_175711924 | 0.26 |
ENST00000444573.1
|
CHN1
|
chimerin 1 |
| chr11_-_8795787 | 0.26 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr10_+_115939008 | 0.25 |
ENST00000369282.1
ENST00000251864.2 ENST00000369281.2 ENST00000422662.1 |
TDRD1
|
tudor domain containing 1 |
| chr5_+_75904918 | 0.25 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr10_-_14596140 | 0.25 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr20_+_20033158 | 0.25 |
ENST00000340348.6
ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26
|
chromosome 20 open reading frame 26 |
| chr17_-_47786375 | 0.25 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr19_-_36019123 | 0.24 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr4_+_159727272 | 0.24 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr19_-_36822551 | 0.24 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr12_-_10321754 | 0.23 |
ENST00000539518.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr17_-_46691990 | 0.23 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr21_-_16374688 | 0.23 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr12_+_21525818 | 0.23 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chrX_+_133941218 | 0.22 |
ENST00000370784.4
ENST00000370785.3 |
FAM122C
|
family with sequence similarity 122C |
| chr15_-_33360342 | 0.22 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr8_+_9953061 | 0.22 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr3_-_196242233 | 0.22 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_+_89829610 | 0.22 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr4_-_170948361 | 0.22 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr11_+_34664014 | 0.22 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr3_-_120400960 | 0.22 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr2_-_175711978 | 0.22 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr2_-_178128149 | 0.22 |
ENST00000423513.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chrX_-_109590174 | 0.22 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr15_+_75639296 | 0.21 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr10_-_16563870 | 0.21 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr12_+_79258444 | 0.21 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr12_+_93963590 | 0.21 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr4_-_186317034 | 0.20 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr14_-_31926623 | 0.20 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr4_-_23891693 | 0.20 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr22_-_41864662 | 0.20 |
ENST00000216252.3
|
PHF5A
|
PHD finger protein 5A |
| chr12_+_32654965 | 0.20 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr3_+_189507523 | 0.19 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr11_-_32452357 | 0.19 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chr4_+_174089951 | 0.19 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr12_-_113841678 | 0.19 |
ENST00000552280.1
ENST00000257549.4 |
SDS
|
serine dehydratase |
| chr5_+_173473662 | 0.19 |
ENST00000519717.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr4_+_170581213 | 0.19 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr11_+_1891380 | 0.19 |
ENST00000429923.1
ENST00000418975.1 ENST00000406638.2 |
LSP1
|
lymphocyte-specific protein 1 |
| chr6_-_28220002 | 0.18 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chrX_+_92929192 | 0.18 |
ENST00000332647.4
|
FAM133A
|
family with sequence similarity 133, member A |
| chr12_+_41221975 | 0.18 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr4_-_146859787 | 0.18 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr19_-_43099070 | 0.18 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr1_+_62439037 | 0.18 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr4_+_128554081 | 0.18 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr1_+_173991633 | 0.17 |
ENST00000424181.1
|
RP11-160H22.3
|
RP11-160H22.3 |
| chr11_-_9088284 | 0.17 |
ENST00000531429.1
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr19_-_46706186 | 0.17 |
ENST00000601263.1
ENST00000598754.1 ENST00000594027.1 ENST00000598518.1 ENST00000599817.1 ENST00000595673.1 |
AC006262.5
|
AC006262.5 |
| chr1_+_203595689 | 0.17 |
ENST00000357681.5
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr6_-_79944336 | 0.17 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr22_+_24105394 | 0.17 |
ENST00000305199.5
ENST00000382821.3 |
C22orf15
|
chromosome 22 open reading frame 15 |
| chr8_+_9953214 | 0.17 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr2_+_145780767 | 0.17 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr3_+_141105705 | 0.17 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr17_-_64188177 | 0.17 |
ENST00000535342.2
|
CEP112
|
centrosomal protein 112kDa |
| chr1_-_146697185 | 0.17 |
ENST00000533174.1
ENST00000254090.4 |
FMO5
|
flavin containing monooxygenase 5 |
| chr12_-_8765446 | 0.17 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr20_-_14318248 | 0.16 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chrX_+_102883887 | 0.16 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr2_+_135596180 | 0.16 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chrX_-_39956656 | 0.15 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr4_-_19458597 | 0.15 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr3_-_196159268 | 0.15 |
ENST00000381887.3
ENST00000535858.1 ENST00000428095.1 ENST00000296328.4 |
UBXN7
|
UBX domain protein 7 |
| chr6_-_88001706 | 0.15 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr15_+_86087267 | 0.15 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr14_-_75083313 | 0.15 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr3_-_97690721 | 0.15 |
ENST00000506099.1
|
MINA
|
MYC induced nuclear antigen |
| chr11_+_74204883 | 0.15 |
ENST00000528481.1
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr3_+_186501979 | 0.15 |
ENST00000498746.1
|
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr8_+_74903580 | 0.14 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr8_-_67579418 | 0.14 |
ENST00000310421.4
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr4_+_169633310 | 0.14 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr15_-_83240783 | 0.14 |
ENST00000568994.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr21_+_43823983 | 0.14 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr4_+_159727222 | 0.14 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr12_-_81331460 | 0.14 |
ENST00000549417.1
|
LIN7A
|
lin-7 homolog A (C. elegans) |
| chr8_+_40010989 | 0.14 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr11_-_108408895 | 0.14 |
ENST00000443411.1
ENST00000533052.1 |
EXPH5
|
exophilin 5 |
| chr10_-_103603523 | 0.14 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr17_+_45608430 | 0.14 |
ENST00000322157.4
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr22_+_41956767 | 0.14 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr19_-_40596828 | 0.14 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chr11_+_76745385 | 0.14 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr15_-_83240507 | 0.14 |
ENST00000564522.1
ENST00000398592.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr10_-_22498950 | 0.13 |
ENST00000422359.2
|
EBLN1
|
endogenous Bornavirus-like nucleoprotein 1 |
| chr19_-_52307357 | 0.13 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr16_+_19125252 | 0.13 |
ENST00000566735.1
ENST00000381440.3 |
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr3_+_57875711 | 0.13 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr9_+_95909309 | 0.13 |
ENST00000366188.2
|
RP11-370F5.4
|
RP11-370F5.4 |
| chr9_-_73483926 | 0.13 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr2_-_3584430 | 0.13 |
ENST00000438482.1
ENST00000422961.1 |
AC108488.4
|
AC108488.4 |
| chr4_+_114214125 | 0.13 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr6_+_74405501 | 0.13 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr4_-_39367949 | 0.13 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr3_-_189839467 | 0.13 |
ENST00000426003.1
|
LEPREL1
|
leprecan-like 1 |
| chr15_+_79165222 | 0.13 |
ENST00000559930.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr5_-_59783882 | 0.13 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr1_+_39796810 | 0.13 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr12_+_79258547 | 0.13 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr15_+_75639951 | 0.13 |
ENST00000564784.1
ENST00000569035.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr5_+_96079240 | 0.13 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr5_+_180467259 | 0.13 |
ENST00000515271.1
|
BTNL9
|
butyrophilin-like 9 |
| chr6_-_91296737 | 0.13 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr17_-_49124230 | 0.13 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr17_-_62208169 | 0.12 |
ENST00000606895.1
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr11_+_65122216 | 0.12 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr1_+_65775204 | 0.12 |
ENST00000371069.4
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr5_-_65018834 | 0.12 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr19_-_40724246 | 0.12 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr1_-_229478714 | 0.12 |
ENST00000284617.2
|
CCSAP
|
centriole, cilia and spindle-associated protein |
| chr1_+_28099683 | 0.12 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr1_+_61547405 | 0.12 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr15_+_59908633 | 0.12 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr5_+_59783540 | 0.12 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr12_+_41221794 | 0.12 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr7_+_114055052 | 0.12 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr8_+_54764346 | 0.12 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr4_-_146859623 | 0.12 |
ENST00000379448.4
ENST00000513320.1 |
ZNF827
|
zinc finger protein 827 |
| chr12_+_510795 | 0.12 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr15_-_83240553 | 0.12 |
ENST00000423133.2
ENST00000398591.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr19_+_6373715 | 0.12 |
ENST00000599849.1
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr13_-_86373536 | 0.12 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr6_-_4135693 | 0.12 |
ENST00000495548.1
ENST00000380125.2 ENST00000465828.1 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr2_-_178128199 | 0.12 |
ENST00000449627.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr12_+_56661461 | 0.12 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr2_+_171036635 | 0.11 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr8_-_68255912 | 0.11 |
ENST00000262215.3
ENST00000519436.1 |
ARFGEF1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr9_+_74764278 | 0.11 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr19_-_9546177 | 0.11 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr6_+_45296048 | 0.11 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr4_-_88312301 | 0.11 |
ENST00000507286.1
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr6_+_31553978 | 0.11 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr20_-_45318230 | 0.11 |
ENST00000372114.3
|
TP53RK
|
TP53 regulating kinase |
| chr11_-_126810521 | 0.11 |
ENST00000530572.1
|
RP11-688I9.4
|
RP11-688I9.4 |
| chr3_-_185826855 | 0.11 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr1_+_167298281 | 0.11 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chrX_-_46187069 | 0.11 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr2_+_152214098 | 0.11 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr11_+_17298342 | 0.11 |
ENST00000530964.1
|
NUCB2
|
nucleobindin 2 |
| chr8_+_132952112 | 0.11 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr15_+_75074915 | 0.11 |
ENST00000567123.1
ENST00000569462.1 |
CSK
|
c-src tyrosine kinase |
| chr4_-_103998439 | 0.11 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr4_-_76957214 | 0.11 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr2_+_187454749 | 0.11 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr7_-_111202511 | 0.11 |
ENST00000452895.1
ENST00000452753.1 ENST00000331762.3 |
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_32259696 | 0.10 |
ENST00000551848.1
|
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr15_+_67841330 | 0.10 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr15_+_42787452 | 0.10 |
ENST00000249647.3
|
SNAP23
|
synaptosomal-associated protein, 23kDa |
| chr20_+_48909240 | 0.10 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr4_-_129209221 | 0.10 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr6_-_33864684 | 0.10 |
ENST00000506222.2
ENST00000533304.1 |
LINC01016
|
long intergenic non-protein coding RNA 1016 |
| chr19_+_17416457 | 0.10 |
ENST00000252602.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr2_+_24150180 | 0.10 |
ENST00000404924.1
|
UBXN2A
|
UBX domain protein 2A |
| chr11_+_111807863 | 0.10 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr3_+_9850199 | 0.10 |
ENST00000452597.1
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3 |
| chr10_-_21186144 | 0.10 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr12_+_11905413 | 0.10 |
ENST00000545027.1
|
ETV6
|
ets variant 6 |
| chrX_+_123097014 | 0.10 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr2_+_110656005 | 0.10 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr5_-_13944652 | 0.10 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.3 | GO:1904640 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.5 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.2 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.4 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.4 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0030969 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.0 | 0.2 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1902868 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:0042866 | L-serine catabolic process(GO:0006565) pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0035566 | negative regulation of icosanoid secretion(GO:0032304) regulation of metanephros size(GO:0035566) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.0 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.8 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.0 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.0 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.2 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.2 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.2 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.2 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |