Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
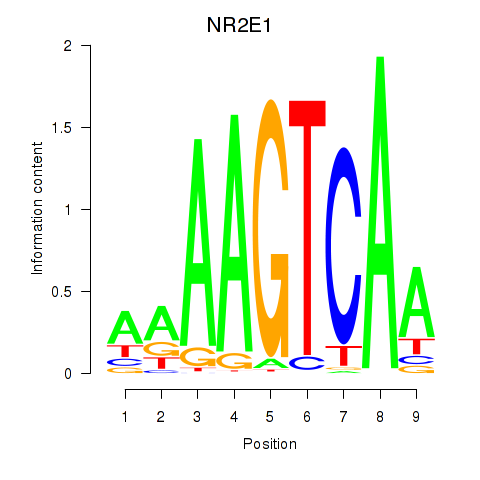
Results for NR2E1
Z-value: 0.20
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | nuclear receptor subfamily 2 group E member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg19_v2_chr6_+_108487245_108487262 | 0.04 | 9.4e-01 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76008706 | 0.15 |
ENST00000562355.1
ENST00000563602.1 |
RP11-44F21.5
|
RP11-44F21.5 |
| chrY_+_15418467 | 0.12 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr2_-_111291587 | 0.09 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr7_+_142829162 | 0.09 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr20_+_62369623 | 0.08 |
ENST00000467211.1
|
RP4-583P15.14
|
RP4-583P15.14 |
| chr6_+_131521120 | 0.08 |
ENST00000537868.1
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr3_-_148939598 | 0.08 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr12_+_86268065 | 0.07 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr3_-_24536453 | 0.07 |
ENST00000453729.2
ENST00000413780.1 |
THRB
|
thyroid hormone receptor, beta |
| chr13_+_95364963 | 0.07 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr3_-_148939835 | 0.07 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr6_+_31916733 | 0.07 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr19_+_35940486 | 0.07 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr12_+_40618873 | 0.07 |
ENST00000298910.7
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr2_+_145780725 | 0.06 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr16_-_15180257 | 0.06 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr3_-_138048653 | 0.06 |
ENST00000460099.1
|
NME9
|
NME/NM23 family member 9 |
| chr14_-_92302784 | 0.06 |
ENST00000340892.5
ENST00000360594.5 |
TC2N
|
tandem C2 domains, nuclear |
| chr12_-_90049878 | 0.06 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr15_-_34502278 | 0.06 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr5_+_125706998 | 0.06 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr5_-_90679145 | 0.05 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr8_-_122653630 | 0.05 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr2_+_29001711 | 0.05 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr3_+_69788576 | 0.05 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr3_+_122044084 | 0.05 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr12_+_75784850 | 0.05 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chrX_-_47863348 | 0.05 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr12_-_90049828 | 0.05 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr6_+_143447322 | 0.05 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chrX_-_106362013 | 0.05 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr6_-_10747802 | 0.04 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr16_-_3068171 | 0.04 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr8_-_17942432 | 0.04 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr19_-_49567124 | 0.04 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr5_+_75699149 | 0.04 |
ENST00000379730.3
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr5_+_67576109 | 0.04 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_-_41039567 | 0.04 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr7_-_105926058 | 0.04 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr12_-_68696652 | 0.04 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr20_-_45980621 | 0.04 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr16_+_22019404 | 0.04 |
ENST00000542527.2
ENST00000569656.1 ENST00000562695.1 |
C16orf52
|
chromosome 16 open reading frame 52 |
| chr10_-_21463116 | 0.04 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr9_-_5304432 | 0.04 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr3_-_4793274 | 0.04 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr17_+_57274914 | 0.04 |
ENST00000582004.1
ENST00000577660.1 |
PRR11
CTD-2510F5.6
|
proline rich 11 Uncharacterized protein |
| chr15_-_34502197 | 0.04 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr9_+_5890802 | 0.04 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr5_-_140013241 | 0.04 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr3_+_138340067 | 0.04 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_-_110883346 | 0.04 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr5_+_75699040 | 0.04 |
ENST00000274364.6
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr5_-_55777586 | 0.04 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr8_-_124279627 | 0.04 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr11_-_126810521 | 0.04 |
ENST00000530572.1
|
RP11-688I9.4
|
RP11-688I9.4 |
| chr7_-_151217166 | 0.04 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr15_+_23255242 | 0.04 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr3_-_101232019 | 0.03 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr8_+_76452097 | 0.03 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chrM_+_10053 | 0.03 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr5_+_56471592 | 0.03 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr14_-_35344093 | 0.03 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr4_+_77177512 | 0.03 |
ENST00000606246.1
|
FAM47E
|
family with sequence similarity 47, member E |
| chr5_-_140013224 | 0.03 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr12_-_31479107 | 0.03 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr11_+_31833939 | 0.03 |
ENST00000530348.1
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr12_+_79258444 | 0.03 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr10_-_29084886 | 0.03 |
ENST00000608061.1
ENST00000443246.2 ENST00000446012.1 |
LINC00837
|
long intergenic non-protein coding RNA 837 |
| chr2_-_190445499 | 0.03 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr9_-_98189055 | 0.03 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr4_-_141348999 | 0.03 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr13_-_49975632 | 0.03 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr12_-_111180644 | 0.03 |
ENST00000551676.1
ENST00000550991.1 ENST00000335007.5 ENST00000340766.5 |
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme |
| chr2_+_198365095 | 0.03 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr3_+_138340049 | 0.03 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_174933899 | 0.03 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr15_-_74374891 | 0.03 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chrX_+_47077680 | 0.03 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr10_-_69991865 | 0.03 |
ENST00000373673.3
|
ATOH7
|
atonal homolog 7 (Drosophila) |
| chr7_-_14026123 | 0.03 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr8_-_16859690 | 0.03 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr7_-_5998714 | 0.03 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr9_-_75567962 | 0.03 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr13_-_31040060 | 0.03 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr11_-_111781454 | 0.03 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr9_-_70465758 | 0.03 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr12_-_31478428 | 0.03 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chrX_-_54824673 | 0.03 |
ENST00000218436.6
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr9_-_119162885 | 0.03 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr12_-_31479045 | 0.03 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr3_-_64673656 | 0.03 |
ENST00000459780.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr20_+_11873141 | 0.03 |
ENST00000422390.1
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr2_+_67624430 | 0.03 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr16_+_21608525 | 0.03 |
ENST00000567404.1
|
METTL9
|
methyltransferase like 9 |
| chr8_+_91013577 | 0.03 |
ENST00000220764.2
|
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr2_-_25194963 | 0.03 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr7_+_116312411 | 0.03 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chrX_+_150866779 | 0.03 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr3_-_179322436 | 0.03 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr9_-_5830768 | 0.03 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr14_+_39734482 | 0.03 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr3_-_119396193 | 0.02 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr4_-_47983519 | 0.02 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr6_+_107077471 | 0.02 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr11_-_111794446 | 0.02 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr6_+_74405501 | 0.02 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr5_-_142780280 | 0.02 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_+_79258547 | 0.02 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr12_+_10366223 | 0.02 |
ENST00000545290.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr12_+_40618764 | 0.02 |
ENST00000343742.2
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr12_+_72061563 | 0.02 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr5_+_173763250 | 0.02 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr8_+_91013676 | 0.02 |
ENST00000519410.1
ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr5_-_135290651 | 0.02 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_+_159141397 | 0.02 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr14_+_31494841 | 0.02 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr2_-_99952769 | 0.02 |
ENST00000409434.1
ENST00000434323.1 ENST00000264255.3 |
TXNDC9
|
thioredoxin domain containing 9 |
| chr11_-_111781554 | 0.02 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr5_-_37371278 | 0.02 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr12_-_75784669 | 0.02 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr4_-_111119804 | 0.02 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr14_+_62164340 | 0.02 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr2_-_87248975 | 0.02 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr6_-_112080256 | 0.02 |
ENST00000462856.2
ENST00000229471.4 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr5_-_9546180 | 0.02 |
ENST00000382496.5
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr12_-_27924209 | 0.02 |
ENST00000381273.3
|
MANSC4
|
MANSC domain containing 4 |
| chrX_-_128657457 | 0.02 |
ENST00000371121.3
ENST00000371123.1 ENST00000371122.4 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr8_-_124408652 | 0.02 |
ENST00000287394.5
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr1_-_27240455 | 0.02 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr1_-_154155675 | 0.02 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr2_+_207630081 | 0.02 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr2_+_3705785 | 0.02 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr14_+_39735411 | 0.02 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr13_+_28813645 | 0.02 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr4_+_22999152 | 0.02 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr6_+_42531798 | 0.02 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr11_-_63439174 | 0.02 |
ENST00000332645.4
|
ATL3
|
atlastin GTPase 3 |
| chr1_+_178995021 | 0.02 |
ENST00000263733.4
|
FAM20B
|
family with sequence similarity 20, member B |
| chr4_+_81187753 | 0.02 |
ENST00000312465.7
ENST00000456523.3 |
FGF5
|
fibroblast growth factor 5 |
| chr12_-_4754339 | 0.02 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr14_+_31494672 | 0.02 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr12_-_10324716 | 0.02 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr1_-_154155595 | 0.02 |
ENST00000328159.4
ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3
|
tropomyosin 3 |
| chr16_+_47495225 | 0.02 |
ENST00000299167.8
ENST00000323584.5 ENST00000563376.1 |
PHKB
|
phosphorylase kinase, beta |
| chr17_-_38956205 | 0.02 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr15_+_31196080 | 0.02 |
ENST00000561607.1
ENST00000565466.1 |
FAN1
|
FANCD2/FANCI-associated nuclease 1 |
| chr2_+_198365122 | 0.02 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr16_-_66952779 | 0.02 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr6_+_10748019 | 0.02 |
ENST00000543878.1
ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L
TMEM14B
|
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr10_+_114709999 | 0.02 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr15_+_83776137 | 0.02 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr12_-_52946923 | 0.02 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr3_-_138048682 | 0.02 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr7_+_116166331 | 0.02 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr16_-_89043377 | 0.02 |
ENST00000436887.2
ENST00000448839.1 ENST00000360302.2 |
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr1_+_90286562 | 0.02 |
ENST00000525774.1
ENST00000337338.5 |
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr5_+_179125907 | 0.02 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr1_-_48866517 | 0.02 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr5_+_9546306 | 0.02 |
ENST00000508179.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding) |
| chr16_+_47495201 | 0.02 |
ENST00000566044.1
ENST00000455779.1 |
PHKB
|
phosphorylase kinase, beta |
| chr6_+_10723148 | 0.02 |
ENST00000541412.1
ENST00000229563.5 |
TMEM14C
|
transmembrane protein 14C |
| chr18_-_6414797 | 0.02 |
ENST00000583809.1
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila) |
| chr3_-_179322416 | 0.02 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chrX_-_138914394 | 0.02 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr2_+_192141611 | 0.02 |
ENST00000392316.1
|
MYO1B
|
myosin IB |
| chr5_-_149516966 | 0.02 |
ENST00000517957.1
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr7_+_106809406 | 0.02 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr3_+_69985734 | 0.02 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr1_+_25944341 | 0.02 |
ENST00000263979.3
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr12_+_120031264 | 0.02 |
ENST00000426426.1
|
TMEM233
|
transmembrane protein 233 |
| chr3_+_69928256 | 0.02 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr10_-_106240032 | 0.02 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr5_+_67588391 | 0.02 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr3_+_113616317 | 0.02 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr5_+_32711419 | 0.02 |
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr2_+_32582086 | 0.02 |
ENST00000421745.2
|
BIRC6
|
baculoviral IAP repeat containing 6 |
| chr6_-_138866823 | 0.02 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr6_-_108582168 | 0.02 |
ENST00000426155.2
|
SNX3
|
sorting nexin 3 |
| chr4_-_13546632 | 0.02 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr12_+_111471828 | 0.02 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr12_+_26348582 | 0.02 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr10_-_94301107 | 0.02 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr6_+_142623758 | 0.02 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr5_+_67588312 | 0.02 |
ENST00000519025.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr17_-_73266616 | 0.02 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr8_+_86121448 | 0.01 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr2_+_110656005 | 0.01 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr12_+_7169887 | 0.01 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr5_-_140013275 | 0.01 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr7_-_38670957 | 0.01 |
ENST00000325590.5
ENST00000428293.2 |
AMPH
|
amphiphysin |
| chr1_-_244013384 | 0.01 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chrX_+_114874727 | 0.01 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr17_+_9479971 | 0.01 |
ENST00000576499.1
|
WDR16
|
WD repeat domain 16 |
| chr7_-_8301682 | 0.01 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr11_-_63439013 | 0.01 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr17_+_9479944 | 0.01 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr7_-_72936531 | 0.01 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr5_-_76935513 | 0.01 |
ENST00000306422.3
|
OTP
|
orthopedia homeobox |
| chr4_-_122085469 | 0.01 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr3_+_14989186 | 0.01 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902823 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.0 | GO:0061193 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.0 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.0 | GO:0008623 | CHRAC(GO:0008623) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.0 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.0 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |