Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
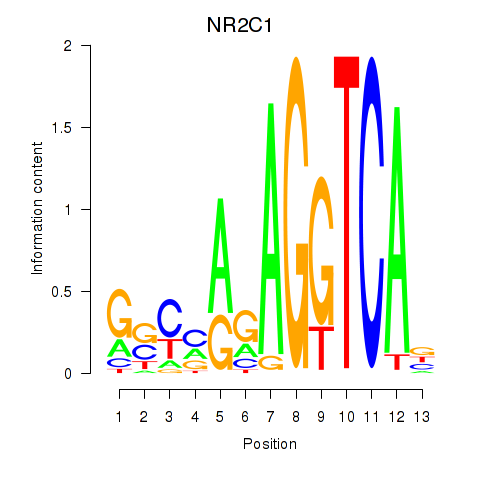
Results for NR2C1
Z-value: 0.58
Transcription factors associated with NR2C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2C1
|
ENSG00000120798.12 | nuclear receptor subfamily 2 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2C1 | hg19_v2_chr12_-_95467267_95467350 | -0.51 | 3.1e-01 | Click! |
Activity profile of NR2C1 motif
Sorted Z-values of NR2C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_12679171 | 0.41 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr2_+_33701684 | 0.27 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_+_6464243 | 0.26 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr19_-_48867171 | 0.25 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr19_+_45116921 | 0.23 |
ENST00000402988.1
|
IGSF23
|
immunoglobulin superfamily, member 23 |
| chr2_-_202316169 | 0.22 |
ENST00000430254.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr1_-_229569834 | 0.21 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr11_-_73687997 | 0.20 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr19_-_51289374 | 0.20 |
ENST00000563228.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr12_-_63544718 | 0.18 |
ENST00000299178.2
|
AVPR1A
|
arginine vasopressin receptor 1A |
| chr12_-_109797249 | 0.18 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr15_+_82555125 | 0.18 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr17_+_13972807 | 0.17 |
ENST00000429152.2
ENST00000261643.3 ENST00000536205.1 ENST00000537334.1 |
COX10
|
cytochrome c oxidase assembly homolog 10 (yeast) |
| chr12_-_6665200 | 0.16 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr19_+_6464502 | 0.15 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr3_+_184055240 | 0.15 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr2_+_74757050 | 0.15 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr21_-_32931290 | 0.15 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr9_+_135037334 | 0.14 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr19_-_48867291 | 0.14 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr22_+_31518938 | 0.14 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr10_+_127661942 | 0.14 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr2_-_202316260 | 0.13 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr18_+_54318893 | 0.13 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr1_+_110577229 | 0.13 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr12_-_22487588 | 0.13 |
ENST00000381424.3
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr3_+_14989186 | 0.13 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_-_55089191 | 0.11 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr12_-_42631529 | 0.10 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr16_-_88752889 | 0.10 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr11_-_45939565 | 0.10 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr12_-_48499826 | 0.10 |
ENST00000551798.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr16_+_83932684 | 0.10 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr5_-_42811986 | 0.10 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_+_44412577 | 0.10 |
ENST00000372343.3
|
IPO13
|
importin 13 |
| chr11_-_26743546 | 0.10 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr3_+_113465866 | 0.10 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr11_-_65429891 | 0.09 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr14_-_73360796 | 0.09 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr8_-_80680078 | 0.09 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr3_+_107318157 | 0.09 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr19_-_46142362 | 0.09 |
ENST00000586770.1
ENST00000591721.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chr17_-_49021974 | 0.09 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr12_+_57854274 | 0.09 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr11_-_64013663 | 0.09 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr9_-_140513231 | 0.09 |
ENST00000371417.3
|
C9orf37
|
chromosome 9 open reading frame 37 |
| chr8_-_145028013 | 0.09 |
ENST00000354958.2
|
PLEC
|
plectin |
| chr15_+_75335604 | 0.09 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr3_+_159557637 | 0.08 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr2_+_219433588 | 0.08 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr19_-_36304201 | 0.08 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr11_+_7595136 | 0.08 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr5_-_42812143 | 0.08 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr15_-_82555000 | 0.08 |
ENST00000557844.1
ENST00000359445.3 ENST00000268206.7 |
EFTUD1
|
elongation factor Tu GTP binding domain containing 1 |
| chr15_-_83474806 | 0.08 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr14_+_105212297 | 0.08 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr15_+_45422178 | 0.08 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr2_+_120125245 | 0.07 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr4_-_99064387 | 0.07 |
ENST00000295268.3
|
STPG2
|
sperm-tail PG-rich repeat containing 2 |
| chr19_+_44100727 | 0.07 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chrX_+_38420783 | 0.07 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr11_+_64073699 | 0.07 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr10_-_73479546 | 0.07 |
ENST00000441508.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr16_-_70729496 | 0.07 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr9_-_34397800 | 0.07 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr19_+_39616410 | 0.07 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr18_+_54318566 | 0.07 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr11_-_45939374 | 0.07 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr17_+_79373540 | 0.07 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr5_+_1801503 | 0.07 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr19_-_36054555 | 0.07 |
ENST00000262623.3
|
ATP4A
|
ATPase, H+/K+ exchanging, alpha polypeptide |
| chr17_-_15903002 | 0.07 |
ENST00000399277.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr2_+_109745655 | 0.07 |
ENST00000418513.1
|
SH3RF3
|
SH3 domain containing ring finger 3 |
| chr17_-_1928621 | 0.06 |
ENST00000331238.6
|
RTN4RL1
|
reticulon 4 receptor-like 1 |
| chr3_-_50605077 | 0.06 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr3_+_183903811 | 0.06 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chrX_-_40036520 | 0.06 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr7_-_15601595 | 0.06 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr7_+_100770328 | 0.06 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr12_-_124873357 | 0.06 |
ENST00000448614.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr14_-_21492251 | 0.06 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr8_-_80942467 | 0.06 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr10_-_103815874 | 0.06 |
ENST00000370033.4
ENST00000311122.5 |
C10orf76
|
chromosome 10 open reading frame 76 |
| chr14_+_74003818 | 0.06 |
ENST00000311148.4
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr13_+_21714711 | 0.06 |
ENST00000607003.1
ENST00000492245.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr4_-_111544254 | 0.06 |
ENST00000306732.3
|
PITX2
|
paired-like homeodomain 2 |
| chr15_+_82555169 | 0.06 |
ENST00000565432.1
ENST00000427381.2 |
FAM154B
|
family with sequence similarity 154, member B |
| chrX_-_19817869 | 0.06 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr3_-_150264272 | 0.06 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr13_+_21714653 | 0.06 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr2_-_201936302 | 0.05 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr17_-_80023659 | 0.05 |
ENST00000578907.1
ENST00000577907.1 ENST00000578176.1 ENST00000582529.1 |
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr12_+_96252706 | 0.05 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr11_-_66206260 | 0.05 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr6_+_10585979 | 0.05 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr20_+_19870167 | 0.05 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr11_+_67798090 | 0.05 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr10_+_24755416 | 0.05 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr1_-_155881156 | 0.05 |
ENST00000539040.1
ENST00000368323.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr16_+_30710462 | 0.05 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr1_+_145727681 | 0.04 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr2_+_219135115 | 0.04 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr4_-_151936416 | 0.04 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr19_-_4066890 | 0.04 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr20_+_30946106 | 0.04 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr13_+_21714913 | 0.04 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr1_+_160097462 | 0.04 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr19_+_48828582 | 0.04 |
ENST00000270221.6
ENST00000596315.1 |
EMP3
|
epithelial membrane protein 3 |
| chr6_+_57037089 | 0.04 |
ENST00000370693.5
|
BAG2
|
BCL2-associated athanogene 2 |
| chr16_+_2255841 | 0.04 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr1_+_26872324 | 0.04 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr9_-_16253112 | 0.04 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr16_+_2255710 | 0.04 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr11_-_64013288 | 0.04 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_-_155880672 | 0.04 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr15_-_65715401 | 0.04 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr1_-_150944411 | 0.03 |
ENST00000368949.4
|
CERS2
|
ceramide synthase 2 |
| chr20_+_327413 | 0.03 |
ENST00000609179.1
|
NRSN2
|
neurensin 2 |
| chr9_-_139839064 | 0.03 |
ENST00000325285.3
ENST00000428398.1 |
FBXW5
|
F-box and WD repeat domain containing 5 |
| chr20_+_44486246 | 0.03 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr11_+_65383227 | 0.03 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr19_+_44100544 | 0.03 |
ENST00000391965.2
ENST00000525771.1 |
ZNF576
|
zinc finger protein 576 |
| chr7_-_72936531 | 0.03 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr19_+_11350278 | 0.03 |
ENST00000252453.8
|
C19orf80
|
chromosome 19 open reading frame 80 |
| chr2_-_42721110 | 0.03 |
ENST00000394973.4
ENST00000306078.1 |
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr19_+_48828788 | 0.03 |
ENST00000594198.1
ENST00000597279.1 ENST00000593437.1 |
EMP3
|
epithelial membrane protein 3 |
| chr2_+_120124497 | 0.03 |
ENST00000355857.3
ENST00000535617.1 ENST00000535757.1 ENST00000409094.1 ENST00000311521.4 |
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr6_+_160148593 | 0.03 |
ENST00000337387.4
|
WTAP
|
Wilms tumor 1 associated protein |
| chr2_+_219433281 | 0.03 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr11_-_67275542 | 0.03 |
ENST00000531506.1
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr6_+_133561725 | 0.03 |
ENST00000452339.2
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr1_-_101491319 | 0.03 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr1_-_173886491 | 0.02 |
ENST00000367698.3
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr2_+_109237717 | 0.02 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_+_67798363 | 0.02 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr7_+_121513143 | 0.02 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr19_+_51293672 | 0.02 |
ENST00000270593.1
ENST00000270594.3 |
ACPT
|
acid phosphatase, testicular |
| chr11_+_67798114 | 0.02 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr7_+_120969045 | 0.02 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chr9_+_33795533 | 0.02 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr17_-_15902903 | 0.02 |
ENST00000486655.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr11_-_64900791 | 0.02 |
ENST00000531018.1
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr13_-_33760216 | 0.02 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr1_+_159750720 | 0.02 |
ENST00000368109.1
ENST00000368108.3 |
DUSP23
|
dual specificity phosphatase 23 |
| chr1_-_917466 | 0.02 |
ENST00000341290.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr15_-_43863695 | 0.02 |
ENST00000439195.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr17_+_80674559 | 0.02 |
ENST00000269373.6
ENST00000535965.1 ENST00000577128.1 ENST00000573158.1 |
FN3KRP
|
fructosamine 3 kinase related protein |
| chr12_-_53729525 | 0.02 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr2_+_33701707 | 0.02 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr22_-_50699701 | 0.02 |
ENST00000395780.1
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr19_-_41256207 | 0.02 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr11_+_94501497 | 0.02 |
ENST00000317829.8
ENST00000317837.9 ENST00000433060.2 |
AMOTL1
|
angiomotin like 1 |
| chr3_-_116164306 | 0.02 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr11_+_131781290 | 0.02 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr4_+_26321284 | 0.01 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_-_50605150 | 0.01 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr19_-_51289436 | 0.01 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr3_+_150264458 | 0.01 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr3_+_179065474 | 0.01 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr1_+_159750776 | 0.01 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr17_+_73997419 | 0.01 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chr9_-_77643189 | 0.01 |
ENST00000376837.3
|
C9orf41
|
chromosome 9 open reading frame 41 |
| chr12_-_54070098 | 0.01 |
ENST00000394349.3
ENST00000549164.1 |
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr22_+_47158578 | 0.01 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr2_-_74757066 | 0.01 |
ENST00000377526.3
|
AUP1
|
ancient ubiquitous protein 1 |
| chr3_+_150264555 | 0.01 |
ENST00000406576.3
ENST00000482093.1 ENST00000273435.5 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr7_-_38948774 | 0.01 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr15_-_43802769 | 0.01 |
ENST00000263801.3
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr6_-_160147925 | 0.01 |
ENST00000535561.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr1_+_165796753 | 0.01 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_+_215255136 | 0.01 |
ENST00000478774.1
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr12_-_54069856 | 0.01 |
ENST00000602871.1
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr2_-_38303218 | 0.01 |
ENST00000407341.1
ENST00000260630.3 |
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr1_+_40713573 | 0.00 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr20_-_57607347 | 0.00 |
ENST00000395663.1
ENST00000395659.1 ENST00000243997.3 |
ATP5E
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr5_+_179125907 | 0.00 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr7_-_72936608 | 0.00 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr15_+_91449971 | 0.00 |
ENST00000557865.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr7_+_143080063 | 0.00 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr17_-_15902951 | 0.00 |
ENST00000472495.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr6_+_54172653 | 0.00 |
ENST00000370869.3
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr8_+_110656344 | 0.00 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
| chr9_-_139137648 | 0.00 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr2_-_219134822 | 0.00 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr19_-_38806560 | 0.00 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr1_-_45140227 | 0.00 |
ENST00000372237.3
|
TMEM53
|
transmembrane protein 53 |
| chr2_+_201936707 | 0.00 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr11_-_72414430 | 0.00 |
ENST00000452383.2
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_-_226374373 | 0.00 |
ENST00000366812.5
|
ACBD3
|
acyl-CoA binding domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2C1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.2 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.2 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of cellular pH reduction(GO:0032849) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.0 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.0 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |