Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
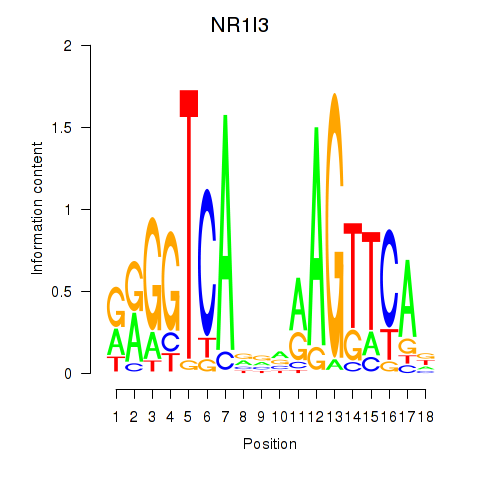
Results for NR1I3
Z-value: 0.28
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.7 | nuclear receptor subfamily 1 group I member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I3 | hg19_v2_chr1_-_161207953_161207975 | -0.52 | 2.9e-01 | Click! |
Activity profile of NR1I3 motif
Sorted Z-values of NR1I3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_153330322 | 0.24 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr1_-_235116495 | 0.22 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr14_-_50506589 | 0.22 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr3_-_71802760 | 0.17 |
ENST00000295612.3
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr12_+_100594557 | 0.16 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr5_-_138780159 | 0.15 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr9_+_134001455 | 0.13 |
ENST00000531584.1
|
NUP214
|
nucleoporin 214kDa |
| chr19_+_2977444 | 0.12 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr17_-_53499310 | 0.11 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr1_+_44401479 | 0.10 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr6_+_36097992 | 0.10 |
ENST00000211287.4
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr5_-_39270725 | 0.09 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr14_+_53173890 | 0.09 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr14_-_22005062 | 0.08 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr14_-_22005197 | 0.08 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr4_-_82136114 | 0.08 |
ENST00000395578.1
ENST00000418486.2 |
PRKG2
|
protein kinase, cGMP-dependent, type II |
| chr2_+_177015950 | 0.07 |
ENST00000306324.3
|
HOXD4
|
homeobox D4 |
| chr2_-_69180083 | 0.07 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr12_-_118810688 | 0.07 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr20_+_5987890 | 0.06 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr13_+_60971080 | 0.06 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr17_-_53499218 | 0.06 |
ENST00000571578.1
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr17_-_62503015 | 0.06 |
ENST00000581806.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr17_-_62502399 | 0.06 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr17_+_32582293 | 0.06 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr1_+_59250815 | 0.05 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr3_+_122103014 | 0.05 |
ENST00000232125.5
ENST00000477892.1 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162, member A |
| chr17_-_62502639 | 0.05 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr14_+_53173910 | 0.05 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr5_+_52856456 | 0.05 |
ENST00000296684.5
ENST00000506765.1 |
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) |
| chr7_-_56160666 | 0.05 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr10_-_73848764 | 0.05 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr7_-_56160625 | 0.05 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr2_-_209118974 | 0.05 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr9_-_80646374 | 0.05 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr8_-_53626974 | 0.05 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr11_-_71781096 | 0.04 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr15_+_43885252 | 0.04 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr10_+_47658234 | 0.04 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr19_-_49140692 | 0.04 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr11_-_61124266 | 0.04 |
ENST00000539890.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr5_-_135528822 | 0.04 |
ENST00000607574.1
|
AC009014.3
|
AC009014.3 |
| chr17_-_26695013 | 0.04 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr1_-_33336414 | 0.03 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr6_+_32407619 | 0.03 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr16_-_66968055 | 0.03 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr6_-_56258892 | 0.03 |
ENST00000370819.1
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chr17_-_8198636 | 0.03 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr1_-_155159748 | 0.03 |
ENST00000473363.2
|
RP11-201K10.3
|
RP11-201K10.3 |
| chr12_-_54779511 | 0.03 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr1_+_145516252 | 0.03 |
ENST00000369306.3
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr11_-_71753188 | 0.03 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr18_-_23670546 | 0.03 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr19_-_6604094 | 0.03 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr17_+_26833250 | 0.03 |
ENST00000577936.1
ENST00000579795.1 |
FOXN1
|
forkhead box N1 |
| chr18_-_23671139 | 0.03 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr1_-_225616515 | 0.03 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr22_+_42372764 | 0.03 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr6_+_71276596 | 0.03 |
ENST00000370474.3
|
C6orf57
|
chromosome 6 open reading frame 57 |
| chr13_-_20806440 | 0.03 |
ENST00000400066.3
ENST00000400065.3 ENST00000356192.6 |
GJB6
|
gap junction protein, beta 6, 30kDa |
| chr9_+_5450503 | 0.03 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr19_+_58920102 | 0.03 |
ENST00000599238.1
ENST00000322834.7 |
ZNF584
|
zinc finger protein 584 |
| chrX_+_110924346 | 0.02 |
ENST00000371979.3
ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr9_-_75567962 | 0.02 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr8_-_143833918 | 0.02 |
ENST00000359228.3
|
LYPD2
|
LY6/PLAUR domain containing 2 |
| chr3_-_58419537 | 0.02 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr14_+_21525981 | 0.02 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr1_-_21625486 | 0.02 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr19_-_44306590 | 0.02 |
ENST00000377950.3
|
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr6_+_96969672 | 0.02 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr8_+_11351876 | 0.02 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr10_+_47657581 | 0.02 |
ENST00000441923.2
|
ANTXRL
|
anthrax toxin receptor-like |
| chr6_+_108487245 | 0.02 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr11_-_82611448 | 0.02 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr19_-_46285646 | 0.02 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr12_-_53729525 | 0.01 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr1_+_82165350 | 0.01 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chrX_+_118602363 | 0.01 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr13_-_114107839 | 0.01 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr14_-_22005018 | 0.01 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr13_+_24883703 | 0.01 |
ENST00000332018.4
|
C1QTNF9
|
C1q and tumor necrosis factor related protein 9 |
| chr1_+_145516560 | 0.01 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr15_+_43985084 | 0.01 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr19_-_48894104 | 0.01 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr1_-_20755140 | 0.01 |
ENST00000418743.1
ENST00000426428.1 |
RP4-749H3.1
|
long intergenic non-protein coding RNA 1141 |
| chr7_-_99381884 | 0.01 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr1_+_160097462 | 0.01 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr5_-_176936817 | 0.01 |
ENST00000502885.1
ENST00000506493.1 |
DOK3
|
docking protein 3 |
| chr1_+_32930647 | 0.01 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr19_-_49140609 | 0.01 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_-_108991812 | 0.01 |
ENST00000392806.3
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chr17_-_33390667 | 0.01 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr22_+_39493268 | 0.01 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr9_+_34179003 | 0.01 |
ENST00000545103.1
ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1
|
ubiquitin associated protein 1 |
| chr4_-_57524061 | 0.00 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chrX_-_50386648 | 0.00 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr17_-_26694979 | 0.00 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr3_-_116163830 | 0.00 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr1_-_32210275 | 0.00 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr1_+_113217043 | 0.00 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr12_-_75601778 | 0.00 |
ENST00000550433.1
ENST00000548513.1 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr1_-_205290865 | 0.00 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr10_+_88414338 | 0.00 |
ENST00000241891.5
ENST00000443292.1 |
OPN4
|
opsin 4 |
| chr16_-_16317321 | 0.00 |
ENST00000205557.7
ENST00000575728.1 |
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chrX_+_69674943 | 0.00 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr19_-_9968816 | 0.00 |
ENST00000590841.1
|
OLFM2
|
olfactomedin 2 |
| chr9_+_75766763 | 0.00 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr2_-_241075706 | 0.00 |
ENST00000607357.1
ENST00000307266.3 |
MYEOV2
|
myeloma overexpressed 2 |
| chr1_+_15250596 | 0.00 |
ENST00000361144.5
|
KAZN
|
kazrin, periplakin interacting protein |
| chr6_-_127780510 | 0.00 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr8_-_52721975 | 0.00 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr15_-_42343388 | 0.00 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |