Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
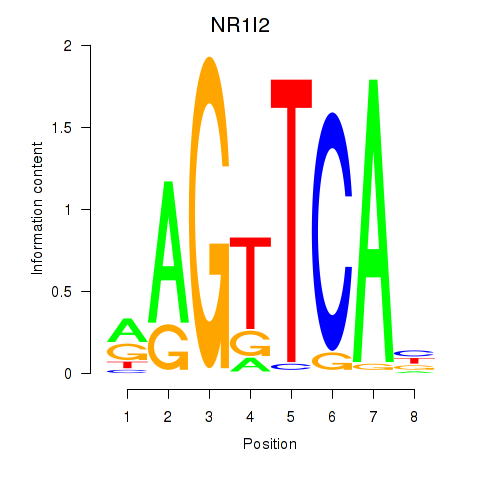
Results for NR1I2
Z-value: 0.75
Transcription factors associated with NR1I2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I2
|
ENSG00000144852.12 | nuclear receptor subfamily 1 group I member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I2 | hg19_v2_chr3_+_119499331_119499331 | -0.69 | 1.3e-01 | Click! |
Activity profile of NR1I2 motif
Sorted Z-values of NR1I2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_12679171 | 0.79 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr9_-_139981121 | 0.57 |
ENST00000596585.1
|
AL807752.1
|
Uncharacterized protein |
| chr1_+_229440129 | 0.49 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr8_-_1922789 | 0.42 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr10_+_77056181 | 0.42 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr3_+_11267691 | 0.40 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr22_+_22676808 | 0.35 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr1_+_15479054 | 0.34 |
ENST00000376014.3
ENST00000451326.2 |
TMEM51
|
transmembrane protein 51 |
| chr16_+_83986827 | 0.33 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr16_-_4164027 | 0.33 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr14_+_73706308 | 0.33 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr19_-_38878632 | 0.31 |
ENST00000586599.1
ENST00000334928.6 ENST00000587676.1 |
GGN
|
gametogenetin |
| chr19_-_44008863 | 0.30 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr3_-_196065248 | 0.30 |
ENST00000446879.1
ENST00000273695.3 |
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr7_-_86848933 | 0.29 |
ENST00000423734.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr4_+_22999152 | 0.29 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr9_-_140115775 | 0.29 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr19_-_36304201 | 0.29 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr17_-_46682321 | 0.29 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr14_-_65769392 | 0.28 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr9_-_118417 | 0.28 |
ENST00000382500.2
|
FOXD4
|
forkhead box D4 |
| chr19_-_39303576 | 0.28 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr16_+_30710462 | 0.27 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr1_+_15479021 | 0.27 |
ENST00000428417.1
|
TMEM51
|
transmembrane protein 51 |
| chr17_-_7531121 | 0.27 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr4_+_71600063 | 0.26 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr6_+_147981838 | 0.26 |
ENST00000427015.1
ENST00000432506.1 |
RP11-307P5.1
|
RP11-307P5.1 |
| chr19_-_3547305 | 0.26 |
ENST00000589063.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr2_+_114256661 | 0.26 |
ENST00000306507.5
|
FOXD4L1
|
forkhead box D4-like 1 |
| chr6_+_73331918 | 0.26 |
ENST00000402622.2
ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr19_+_10947251 | 0.26 |
ENST00000592854.1
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr16_-_4466565 | 0.25 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr11_+_117073850 | 0.25 |
ENST00000529622.1
|
TAGLN
|
transgelin |
| chr8_-_9009079 | 0.25 |
ENST00000519699.1
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr16_-_88752889 | 0.25 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr16_+_28875126 | 0.24 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr5_+_139055055 | 0.24 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr5_-_154230130 | 0.24 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr16_+_28874345 | 0.24 |
ENST00000566209.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr15_+_96876340 | 0.24 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_-_6665200 | 0.23 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr9_+_21440440 | 0.23 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr1_+_43291220 | 0.23 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr16_-_15950868 | 0.22 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr6_-_32977345 | 0.22 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr13_+_24844857 | 0.22 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr3_+_193853927 | 0.22 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr11_+_118478313 | 0.22 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr13_+_24844819 | 0.21 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr21_-_44751903 | 0.21 |
ENST00000450205.1
|
LINC00322
|
long intergenic non-protein coding RNA 322 |
| chr11_+_66624527 | 0.21 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr6_+_12007963 | 0.21 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr17_-_33448468 | 0.21 |
ENST00000591723.1
ENST00000593039.1 ENST00000587405.1 |
RAD51L3-RFFL
RAD51D
|
Uncharacterized protein RAD51 paralog D |
| chr3_-_9994021 | 0.21 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr12_+_117176113 | 0.21 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2 |
| chr7_-_150777920 | 0.21 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr6_-_34664612 | 0.21 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr6_-_152623231 | 0.21 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr19_-_43690674 | 0.21 |
ENST00000342951.6
ENST00000366175.3 |
PSG5
|
pregnancy specific beta-1-glycoprotein 5 |
| chr4_+_71600144 | 0.21 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_-_178969403 | 0.21 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr16_+_58498194 | 0.20 |
ENST00000561779.1
ENST00000565430.1 ENST00000567063.1 ENST00000566041.1 |
NDRG4
|
NDRG family member 4 |
| chr19_-_51289374 | 0.20 |
ENST00000563228.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr1_-_55089191 | 0.20 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr12_+_120875910 | 0.20 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr2_-_31440377 | 0.20 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr16_-_58328923 | 0.20 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr1_+_12538594 | 0.20 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr16_-_28937027 | 0.19 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr10_-_103347883 | 0.19 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr2_+_30455016 | 0.19 |
ENST00000401506.1
ENST00000407930.2 |
LBH
|
limb bud and heart development |
| chr20_+_62492566 | 0.19 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr3_+_8543393 | 0.19 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr2_+_109223595 | 0.19 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_-_158757895 | 0.19 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr11_-_134095335 | 0.18 |
ENST00000534227.1
ENST00000532445.1 |
NCAPD3
|
non-SMC condensin II complex, subunit D3 |
| chr18_+_3466248 | 0.18 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr11_+_107461948 | 0.18 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chrX_-_30327495 | 0.18 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr16_+_84209539 | 0.18 |
ENST00000569735.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr7_-_150777949 | 0.18 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr7_-_56174161 | 0.18 |
ENST00000395422.3
|
CHCHD2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr8_+_99956662 | 0.18 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr14_-_95236551 | 0.18 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr20_+_3776371 | 0.18 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr2_+_242127924 | 0.18 |
ENST00000402530.3
ENST00000274979.8 ENST00000402430.3 |
ANO7
|
anoctamin 7 |
| chr3_+_8543533 | 0.17 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr22_+_50354104 | 0.17 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr5_+_139055021 | 0.17 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr19_+_39616410 | 0.17 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr14_+_23340822 | 0.17 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr7_-_149158187 | 0.17 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr5_+_1801503 | 0.17 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr15_+_42696992 | 0.17 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chr6_+_158733692 | 0.17 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr3_-_185826286 | 0.17 |
ENST00000537818.1
ENST00000422039.1 ENST00000434744.1 |
ETV5
|
ets variant 5 |
| chr12_-_124873357 | 0.16 |
ENST00000448614.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr16_+_29991673 | 0.16 |
ENST00000416441.2
|
TAOK2
|
TAO kinase 2 |
| chr20_+_19870167 | 0.16 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr10_+_24755416 | 0.16 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr14_+_64565442 | 0.16 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr17_-_4852332 | 0.16 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr1_+_156589051 | 0.16 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr2_+_109237717 | 0.16 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr6_+_43738444 | 0.16 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr1_+_223101757 | 0.16 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr8_+_99956759 | 0.16 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr1_+_156589198 | 0.16 |
ENST00000456112.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr1_-_151299842 | 0.16 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr22_+_46546406 | 0.16 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr7_-_131241361 | 0.16 |
ENST00000378555.3
ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL
|
podocalyxin-like |
| chr2_-_75426183 | 0.16 |
ENST00000409848.3
|
TACR1
|
tachykinin receptor 1 |
| chr2_-_200335983 | 0.16 |
ENST00000457245.1
|
SATB2
|
SATB homeobox 2 |
| chr4_-_152682129 | 0.16 |
ENST00000512306.1
ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112
|
PET112 homolog (yeast) |
| chr4_-_110723134 | 0.16 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr2_-_69870835 | 0.16 |
ENST00000409085.4
ENST00000406297.3 |
AAK1
|
AP2 associated kinase 1 |
| chr21_+_43823983 | 0.16 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr3_+_8543561 | 0.16 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr19_+_9251052 | 0.15 |
ENST00000247956.6
ENST00000360385.3 |
ZNF317
|
zinc finger protein 317 |
| chr11_+_62649158 | 0.15 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chrX_+_49832231 | 0.15 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr9_-_14314066 | 0.15 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chr6_-_53013620 | 0.15 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr16_-_2770216 | 0.15 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr3_-_196065374 | 0.15 |
ENST00000454715.1
|
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr22_-_43567750 | 0.15 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr7_-_105332084 | 0.15 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr2_+_30454390 | 0.15 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr19_+_36236514 | 0.15 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr2_+_233734994 | 0.15 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr13_-_95364389 | 0.15 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr14_-_76447336 | 0.15 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor, beta 3 |
| chr4_+_113970772 | 0.15 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr1_-_54879140 | 0.15 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr15_-_89438742 | 0.15 |
ENST00000562281.1
ENST00000562889.1 ENST00000359595.3 |
HAPLN3
|
hyaluronan and proteoglycan link protein 3 |
| chr6_-_138893661 | 0.15 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr4_-_120243545 | 0.14 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr16_+_1832902 | 0.14 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr1_-_229569834 | 0.14 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr9_+_35829208 | 0.14 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr1_-_205091115 | 0.14 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr9_-_34620440 | 0.14 |
ENST00000421919.1
ENST00000378911.3 ENST00000477738.2 ENST00000341694.2 ENST00000259632.7 ENST00000378913.2 ENST00000378916.4 ENST00000447983.2 |
DCTN3
|
dynactin 3 (p22) |
| chr19_+_11039391 | 0.14 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr9_-_73483926 | 0.14 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr7_+_97910962 | 0.14 |
ENST00000539286.1
|
BRI3
|
brain protein I3 |
| chr1_+_201979645 | 0.14 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr10_-_76995675 | 0.14 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr15_+_59730348 | 0.14 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr2_-_69870747 | 0.14 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr16_-_79804435 | 0.14 |
ENST00000562921.1
ENST00000566729.1 |
RP11-345M22.2
|
RP11-345M22.2 |
| chr1_+_36621174 | 0.14 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr1_-_53387386 | 0.14 |
ENST00000467988.1
ENST00000358358.5 ENST00000371522.4 |
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr4_-_110723194 | 0.14 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr3_-_123123407 | 0.14 |
ENST00000466617.1
|
ADCY5
|
adenylate cyclase 5 |
| chr10_-_105677427 | 0.14 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr12_-_52585765 | 0.14 |
ENST00000313234.5
ENST00000394815.2 |
KRT80
|
keratin 80 |
| chr14_+_100532771 | 0.14 |
ENST00000557153.1
|
EVL
|
Enah/Vasp-like |
| chr8_+_21911054 | 0.13 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr1_+_145209092 | 0.13 |
ENST00000362074.6
ENST00000344859.3 |
NOTCH2NL
|
notch 2 N-terminal like |
| chr2_-_74779744 | 0.13 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr2_-_86790593 | 0.13 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr15_-_43212836 | 0.13 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr19_-_45657028 | 0.13 |
ENST00000429338.1
ENST00000589776.1 |
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr9_+_139981513 | 0.13 |
ENST00000535144.1
ENST00000542372.1 |
MAN1B1
|
mannosidase, alpha, class 1B, member 1 |
| chr1_-_144994840 | 0.13 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr19_-_10946871 | 0.12 |
ENST00000589638.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr17_-_14683517 | 0.12 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr1_+_201979743 | 0.12 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr5_-_158526756 | 0.12 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr3_-_114343039 | 0.12 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr13_-_30424821 | 0.12 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr12_+_57854274 | 0.12 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr1_+_110754094 | 0.12 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr19_-_12845467 | 0.12 |
ENST00000592273.1
ENST00000588213.1 |
C19orf43
|
chromosome 19 open reading frame 43 |
| chr22_+_25003568 | 0.12 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr11_+_62648336 | 0.12 |
ENST00000338663.7
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr1_+_24646263 | 0.12 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_149814478 | 0.12 |
ENST00000369161.3
|
HIST2H2AA3
|
histone cluster 2, H2aa3 |
| chr1_+_32379174 | 0.12 |
ENST00000391369.1
|
AL136115.1
|
HCG2032337; PRO1848; Uncharacterized protein |
| chr9_-_130966497 | 0.12 |
ENST00000393608.1
ENST00000372948.3 |
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr6_-_27782548 | 0.12 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr19_-_12845550 | 0.12 |
ENST00000242784.4
|
C19orf43
|
chromosome 19 open reading frame 43 |
| chr7_-_111424462 | 0.12 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr15_+_96875657 | 0.12 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr3_-_117716418 | 0.12 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr18_+_29027696 | 0.11 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr17_-_2169425 | 0.11 |
ENST00000570606.1
ENST00000354901.4 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr6_+_37897735 | 0.11 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr3_+_14989186 | 0.11 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr8_-_29120580 | 0.11 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr2_+_97426631 | 0.11 |
ENST00000377075.2
|
CNNM4
|
cyclin M4 |
| chr17_-_9929581 | 0.11 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr11_-_111794446 | 0.11 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr3_+_69985734 | 0.11 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr3_+_183894737 | 0.11 |
ENST00000432591.1
ENST00000431779.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr9_+_139981379 | 0.11 |
ENST00000371589.4
|
MAN1B1
|
mannosidase, alpha, class 1B, member 1 |
| chr6_+_45296048 | 0.11 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr5_+_73109339 | 0.11 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr8_+_86999516 | 0.11 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr14_-_73493825 | 0.11 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.4 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.2 | GO:0045632 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.2 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.3 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.2 | GO:1903570 | coronary vein morphogenesis(GO:0003169) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.3 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.2 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.4 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.1 | GO:1903762 | regulation of heart looping(GO:1901207) positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.0 | GO:2001113 | negative regulation of hepatocyte growth factor receptor signaling pathway(GO:1902203) regulation of cellular response to hepatocyte growth factor stimulus(GO:2001112) negative regulation of cellular response to hepatocyte growth factor stimulus(GO:2001113) |
| 0.0 | 0.5 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0032982 | myosin filament(GO:0032982) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.0 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |