Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
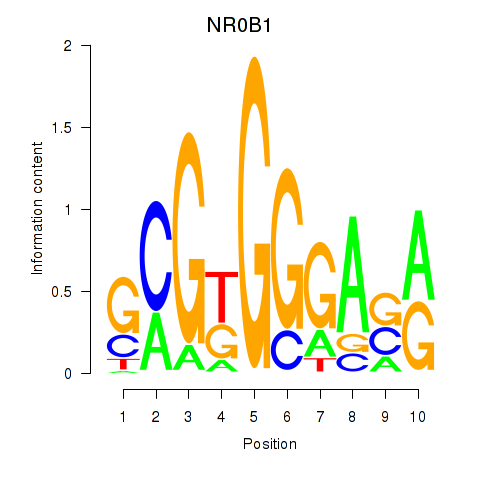
Results for NR0B1
Z-value: 0.90
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | nuclear receptor subfamily 0 group B member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR0B1 | hg19_v2_chrX_-_30326445_30326605 | 0.45 | 3.7e-01 | Click! |
Activity profile of NR0B1 motif
Sorted Z-values of NR0B1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_4845379 | 1.17 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr7_+_26438187 | 0.76 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr3_-_64431146 | 0.56 |
ENST00000569824.1
ENST00000485770.1 |
PRICKLE2
RP11-14D22.2
|
prickle homolog 2 (Drosophila) RP11-14D22.2 |
| chr4_-_90758227 | 0.52 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr5_+_111496631 | 0.49 |
ENST00000508590.1
|
EPB41L4A-AS1
|
EPB41L4A antisense RNA 1 |
| chr11_+_130184888 | 0.44 |
ENST00000602376.1
ENST00000532116.3 ENST00000602310.1 |
RP11-121M22.1
|
RP11-121M22.1 |
| chr17_+_29421900 | 0.43 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr3_-_176915036 | 0.43 |
ENST00000427349.1
ENST00000352800.5 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr20_-_18038521 | 0.42 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chr2_+_105471969 | 0.40 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr2_-_165698322 | 0.39 |
ENST00000444537.1
ENST00000414843.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr9_-_134615326 | 0.37 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr1_+_203765437 | 0.37 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr16_+_67280799 | 0.35 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr3_-_113415441 | 0.31 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr6_+_56954867 | 0.29 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chrX_-_20284733 | 0.28 |
ENST00000438357.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr12_-_80084333 | 0.28 |
ENST00000552637.1
|
PAWR
|
PRKC, apoptosis, WT1, regulator |
| chr19_-_49622348 | 0.27 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr6_-_111804393 | 0.26 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr11_+_117015024 | 0.25 |
ENST00000530272.1
|
PAFAH1B2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) |
| chr16_-_18937726 | 0.25 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr8_+_100025476 | 0.25 |
ENST00000355155.1
ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast) |
| chr7_-_48068643 | 0.24 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr17_+_29421987 | 0.24 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr6_-_110500826 | 0.24 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr14_-_61190754 | 0.23 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr14_+_59104741 | 0.23 |
ENST00000395153.3
ENST00000335867.4 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr1_-_159915386 | 0.23 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr1_+_2477831 | 0.23 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12 |
| chr19_+_35842445 | 0.22 |
ENST00000246553.2
|
FFAR1
|
free fatty acid receptor 1 |
| chr2_-_159313214 | 0.22 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr1_-_223537401 | 0.22 |
ENST00000343846.3
ENST00000454695.2 ENST00000484758.2 |
SUSD4
|
sushi domain containing 4 |
| chr10_-_123687431 | 0.22 |
ENST00000423243.1
|
ATE1
|
arginyltransferase 1 |
| chr6_-_111804905 | 0.21 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr17_-_65241281 | 0.21 |
ENST00000358691.5
ENST00000580168.1 |
HELZ
|
helicase with zinc finger |
| chr11_-_117667806 | 0.21 |
ENST00000527706.1
ENST00000321322.6 |
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr9_+_128509624 | 0.21 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr8_-_144679264 | 0.20 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr5_-_142784003 | 0.20 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr19_+_56111680 | 0.20 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr11_+_64053311 | 0.20 |
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr10_-_17659234 | 0.20 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr12_-_99288536 | 0.20 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr17_-_27181990 | 0.20 |
ENST00000583953.1
|
FAM222B
|
family with sequence similarity 222, member B |
| chr8_+_42396712 | 0.20 |
ENST00000518574.1
ENST00000417410.2 ENST00000414154.2 |
SMIM19
|
small integral membrane protein 19 |
| chr5_-_142783694 | 0.20 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_+_50355647 | 0.19 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr12_+_107168418 | 0.19 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr1_-_46598371 | 0.19 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr2_+_209131059 | 0.17 |
ENST00000422495.1
ENST00000452564.1 |
PIKFYVE
|
phosphoinositide kinase, FYVE finger containing |
| chr6_-_48036363 | 0.17 |
ENST00000543600.1
ENST00000398738.2 ENST00000339488.4 |
PTCHD4
|
patched domain containing 4 |
| chr1_+_235492300 | 0.17 |
ENST00000476121.1
ENST00000497327.1 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr12_+_49372251 | 0.17 |
ENST00000293549.3
|
WNT1
|
wingless-type MMTV integration site family, member 1 |
| chr19_+_58694742 | 0.16 |
ENST00000597528.1
ENST00000594839.1 |
ZNF274
|
zinc finger protein 274 |
| chr4_+_38665810 | 0.16 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr1_+_211433275 | 0.16 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chrX_-_112084043 | 0.16 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr11_-_61101247 | 0.16 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chr12_+_65672702 | 0.16 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr22_+_29876197 | 0.16 |
ENST00000310624.6
|
NEFH
|
neurofilament, heavy polypeptide |
| chr17_+_45608614 | 0.15 |
ENST00000544660.1
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr19_-_4722780 | 0.15 |
ENST00000600621.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr16_+_3014217 | 0.15 |
ENST00000572045.1
|
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr12_-_58131931 | 0.15 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr17_-_4890919 | 0.15 |
ENST00000572543.1
ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr3_+_88108381 | 0.14 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr21_-_32931290 | 0.14 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr19_-_49576198 | 0.14 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr19_-_15090488 | 0.14 |
ENST00000594383.1
ENST00000598504.1 ENST00000597262.1 |
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr2_+_220306238 | 0.14 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr10_-_13570533 | 0.13 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr10_-_128077024 | 0.13 |
ENST00000368679.4
ENST00000368676.4 ENST00000448723.1 |
ADAM12
|
ADAM metallopeptidase domain 12 |
| chr4_+_83351791 | 0.13 |
ENST00000509635.1
|
ENOPH1
|
enolase-phosphatase 1 |
| chr20_+_49348081 | 0.13 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr7_+_2559399 | 0.13 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr15_-_28957469 | 0.13 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr11_+_57435441 | 0.13 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr4_-_151936416 | 0.13 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr11_-_12030681 | 0.13 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr1_-_236030216 | 0.13 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr6_+_20403997 | 0.13 |
ENST00000535432.1
|
E2F3
|
E2F transcription factor 3 |
| chr8_-_100025238 | 0.12 |
ENST00000521696.1
|
RP11-410L14.2
|
RP11-410L14.2 |
| chr2_+_170683979 | 0.12 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr9_-_133814527 | 0.12 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr12_-_39837192 | 0.12 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr1_-_247275719 | 0.12 |
ENST00000408893.2
|
C1orf229
|
chromosome 1 open reading frame 229 |
| chr16_+_89894875 | 0.12 |
ENST00000393062.2
|
SPIRE2
|
spire-type actin nucleation factor 2 |
| chr21_-_34100244 | 0.12 |
ENST00000382491.3
ENST00000357345.3 ENST00000429236.1 |
SYNJ1
|
synaptojanin 1 |
| chr3_-_176914963 | 0.12 |
ENST00000450267.1
ENST00000431674.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr6_+_7107830 | 0.12 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr17_-_28618867 | 0.12 |
ENST00000394819.3
ENST00000577623.1 |
BLMH
|
bleomycin hydrolase |
| chr11_-_65625014 | 0.11 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr1_+_52608046 | 0.11 |
ENST00000357206.2
ENST00000287727.3 |
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr8_-_27469383 | 0.11 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr19_+_48673949 | 0.11 |
ENST00000328759.7
|
C19orf68
|
chromosome 19 open reading frame 68 |
| chr3_-_33260707 | 0.11 |
ENST00000309558.3
|
SUSD5
|
sushi domain containing 5 |
| chr1_-_46598284 | 0.11 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr10_+_38299546 | 0.11 |
ENST00000374618.3
ENST00000432900.2 ENST00000458705.2 ENST00000469037.2 |
ZNF33A
|
zinc finger protein 33A |
| chr7_-_8301682 | 0.10 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chrX_+_133507389 | 0.10 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr3_-_123168551 | 0.10 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr11_+_57509020 | 0.10 |
ENST00000388857.4
ENST00000528798.1 |
C11orf31
|
chromosome 11 open reading frame 31 |
| chr17_-_27332931 | 0.10 |
ENST00000442608.3
ENST00000335960.6 |
SEZ6
|
seizure related 6 homolog (mouse) |
| chr9_-_110251836 | 0.10 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr3_+_128720424 | 0.10 |
ENST00000480450.1
ENST00000436022.2 |
EFCC1
|
EF-hand and coiled-coil domain containing 1 |
| chr12_+_56473910 | 0.10 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr3_+_170075436 | 0.10 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr19_-_52097613 | 0.10 |
ENST00000301439.3
|
AC018755.1
|
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chr6_-_139308777 | 0.10 |
ENST00000529597.1
ENST00000415951.2 ENST00000367663.4 ENST00000409812.2 |
REPS1
|
RALBP1 associated Eps domain containing 1 |
| chr3_-_11623804 | 0.10 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr20_+_57466629 | 0.10 |
ENST00000371081.1
ENST00000338783.6 |
GNAS
|
GNAS complex locus |
| chr17_+_40811283 | 0.10 |
ENST00000251412.7
|
TUBG2
|
tubulin, gamma 2 |
| chr12_-_112037136 | 0.09 |
ENST00000608853.1
|
ATXN2
|
ataxin 2 |
| chr6_-_29595779 | 0.09 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chrX_-_114468605 | 0.09 |
ENST00000538422.1
ENST00000317135.8 |
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr8_-_142011036 | 0.09 |
ENST00000520892.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr16_+_14280564 | 0.09 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr2_+_201171577 | 0.09 |
ENST00000409397.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr20_-_35807741 | 0.09 |
ENST00000434295.1
ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr19_-_40324255 | 0.09 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr15_-_51914810 | 0.09 |
ENST00000543779.2
ENST00000449909.3 |
DMXL2
|
Dmx-like 2 |
| chr8_-_144679602 | 0.09 |
ENST00000526710.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrX_+_133930798 | 0.09 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr4_-_90758118 | 0.09 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr21_-_34100341 | 0.09 |
ENST00000382499.2
ENST00000433931.2 |
SYNJ1
|
synaptojanin 1 |
| chr4_-_151936865 | 0.09 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr3_-_52567792 | 0.09 |
ENST00000307092.4
ENST00000422318.2 ENST00000459839.1 |
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr2_-_43823093 | 0.09 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr7_+_89841144 | 0.09 |
ENST00000394622.2
ENST00000394632.1 ENST00000426158.1 ENST00000394621.2 ENST00000402625.2 |
STEAP2
|
STEAP family member 2, metalloreductase |
| chr17_+_46018872 | 0.09 |
ENST00000583599.1
ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr10_-_123687497 | 0.09 |
ENST00000369040.3
ENST00000224652.6 ENST00000369043.3 |
ATE1
|
arginyltransferase 1 |
| chr5_+_156887027 | 0.09 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr6_+_33388013 | 0.09 |
ENST00000449372.2
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr4_-_146101304 | 0.08 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chrX_-_131352152 | 0.08 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr3_+_111578583 | 0.08 |
ENST00000478922.1
ENST00000477695.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr15_+_74833518 | 0.08 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr17_+_7155343 | 0.08 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr10_+_5726764 | 0.08 |
ENST00000328090.5
ENST00000496681.1 |
FAM208B
|
family with sequence similarity 208, member B |
| chr19_+_55795493 | 0.08 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr1_+_183605222 | 0.08 |
ENST00000536277.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr5_+_138609441 | 0.08 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr21_+_40181520 | 0.08 |
ENST00000456966.1
|
ETS2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr2_-_43823119 | 0.08 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr7_-_22862406 | 0.08 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr8_-_28347737 | 0.08 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr2_+_54198210 | 0.08 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr5_+_138609782 | 0.08 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr12_+_107168342 | 0.08 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr20_+_35202909 | 0.08 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr1_+_180601139 | 0.08 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr12_+_51985001 | 0.08 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr19_-_36523709 | 0.08 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr13_-_110438914 | 0.08 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr3_-_45957088 | 0.08 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr6_-_112575687 | 0.08 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr6_+_37787704 | 0.07 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr12_+_41086297 | 0.07 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr5_+_67511524 | 0.07 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_+_69864129 | 0.07 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr2_+_170683942 | 0.07 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr17_-_40761375 | 0.07 |
ENST00000543197.1
ENST00000309428.5 |
FAM134C
|
family with sequence similarity 134, member C |
| chr12_+_113860160 | 0.07 |
ENST00000553248.1
ENST00000345635.4 ENST00000547802.1 |
SDSL
|
serine dehydratase-like |
| chr5_-_159739532 | 0.07 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr6_-_160210604 | 0.07 |
ENST00000420894.2
ENST00000539756.1 ENST00000544255.1 |
TCP1
|
t-complex 1 |
| chr4_-_2758015 | 0.07 |
ENST00000510267.1
ENST00000503235.1 ENST00000315423.7 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr10_-_17659357 | 0.07 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr2_+_234580499 | 0.07 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr12_+_27677085 | 0.07 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr11_-_119993979 | 0.07 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr22_-_31885727 | 0.07 |
ENST00000330125.5
ENST00000344710.5 ENST00000397518.1 |
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr11_-_92930556 | 0.07 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr2_+_204193149 | 0.07 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr19_+_41036371 | 0.07 |
ENST00000392023.1
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr6_-_112575838 | 0.07 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr14_-_21905424 | 0.07 |
ENST00000553622.1
|
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr8_+_38644778 | 0.06 |
ENST00000276520.8
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr3_+_171758344 | 0.06 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr11_-_64512273 | 0.06 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr1_+_26146397 | 0.06 |
ENST00000374303.2
ENST00000533762.1 ENST00000529116.1 ENST00000474295.1 ENST00000488327.2 ENST00000472643.1 ENST00000526894.1 ENST00000524618.1 ENST00000374307.5 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr2_+_217277137 | 0.06 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr1_+_226250379 | 0.06 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr16_+_14280742 | 0.06 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr11_+_32914579 | 0.06 |
ENST00000399302.2
|
QSER1
|
glutamine and serine rich 1 |
| chr21_-_15755446 | 0.06 |
ENST00000544452.1
ENST00000285667.3 |
HSPA13
|
heat shock protein 70kDa family, member 13 |
| chr2_+_234580525 | 0.06 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_-_87379785 | 0.06 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr1_+_53527854 | 0.06 |
ENST00000371500.3
ENST00000395871.2 ENST00000312553.5 |
PODN
|
podocan |
| chr13_+_47127293 | 0.06 |
ENST00000311191.6
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr12_-_58240470 | 0.06 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr11_+_61447845 | 0.06 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr17_-_7155775 | 0.06 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr3_-_47517302 | 0.06 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr10_-_112064665 | 0.06 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr2_+_204192942 | 0.06 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr7_-_8301768 | 0.06 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr19_+_532049 | 0.06 |
ENST00000606136.1
|
CDC34
|
cell division cycle 34 |
| chr19_+_49660997 | 0.06 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr17_-_27333163 | 0.06 |
ENST00000360295.9
|
SEZ6
|
seizure related 6 homolog (mouse) |
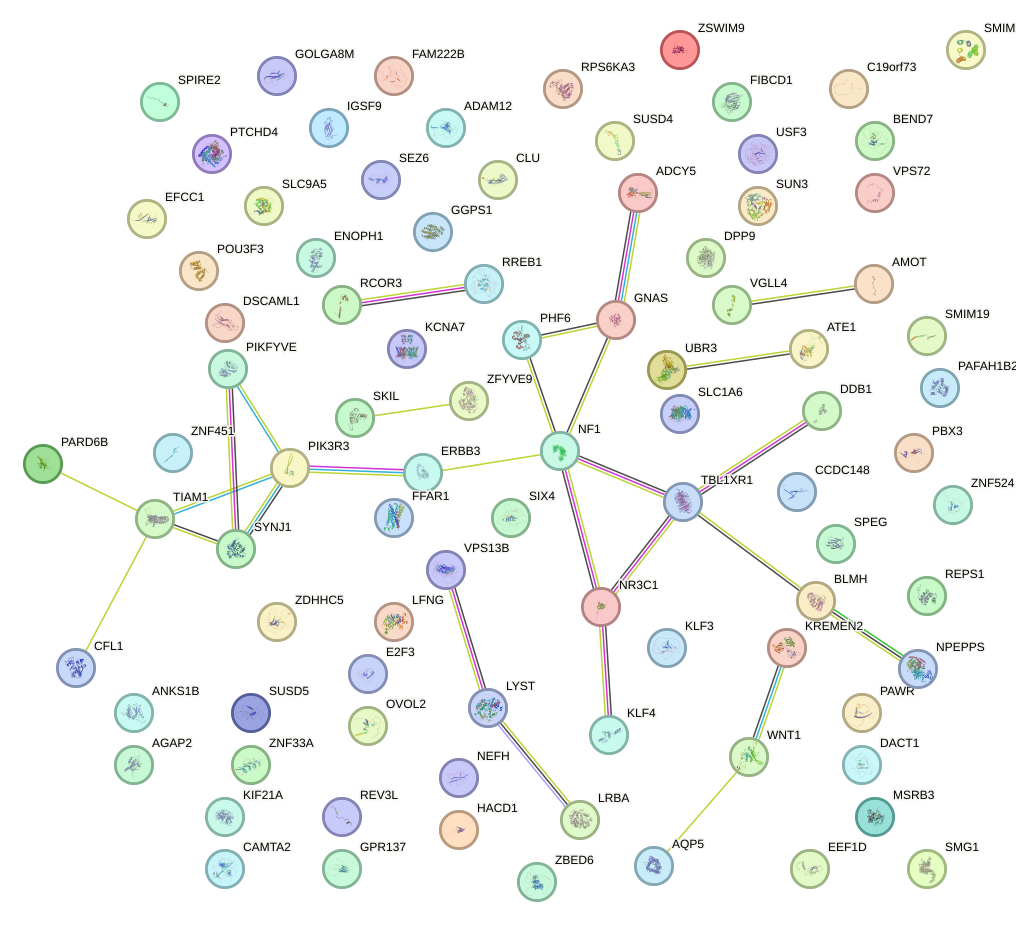
Network of associatons between targets according to the STRING database.
First level regulatory network of NR0B1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.6 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.3 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.5 | GO:0072218 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.1 | 0.3 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.4 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.2 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.2 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.2 | GO:0061054 | Spemann organizer formation(GO:0060061) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) |
| 0.1 | 0.2 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.2 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:1904995 | regulation of muscle hyperplasia(GO:0014738) negative regulation of muscle hyperplasia(GO:0014740) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.2 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0098712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) L-glutamate import across plasma membrane(GO:0098712) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0070632 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.1 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.0 | 0.1 | GO:2000771 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.1 | 0.4 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |