Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
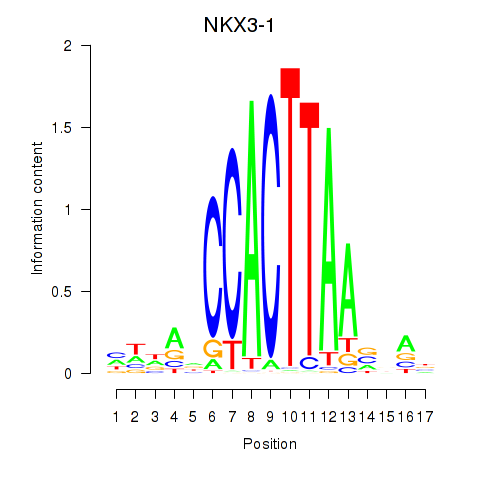
Results for NKX3-1
Z-value: 0.47
Transcription factors associated with NKX3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-1
|
ENSG00000167034.9 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-1 | hg19_v2_chr8_-_23540402_23540466 | 0.01 | 9.8e-01 | Click! |
Activity profile of NKX3-1 motif
Sorted Z-values of NKX3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_149785236 | 0.90 |
ENST00000331491.1
|
HIST2H3D
|
histone cluster 2, H3d |
| chr10_-_99185798 | 0.39 |
ENST00000439965.2
|
AL355490.1
|
LOC644215 protein; Uncharacterized protein |
| chr2_-_37374876 | 0.35 |
ENST00000405334.1
|
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr16_+_47496023 | 0.34 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr12_+_113354341 | 0.33 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr4_+_17579110 | 0.29 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr5_+_68485433 | 0.26 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr12_-_118628315 | 0.25 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr6_+_42928485 | 0.25 |
ENST00000372808.3
|
GNMT
|
glycine N-methyltransferase |
| chr6_+_26217159 | 0.23 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr21_+_35736302 | 0.23 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr12_+_109569155 | 0.22 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr1_-_89488510 | 0.20 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr7_-_33080506 | 0.20 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr14_+_67656110 | 0.20 |
ENST00000524532.1
ENST00000530728.1 |
FAM71D
|
family with sequence similarity 71, member D |
| chr1_+_196621156 | 0.19 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr18_+_61445007 | 0.19 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr15_-_31521567 | 0.18 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr6_+_149887377 | 0.17 |
ENST00000367419.5
|
GINM1
|
glycoprotein integral membrane 1 |
| chr15_+_63682335 | 0.17 |
ENST00000559379.1
ENST00000559821.1 |
RP11-321G12.1
|
RP11-321G12.1 |
| chr14_+_61449197 | 0.17 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr19_-_12708113 | 0.17 |
ENST00000440366.1
|
ZNF490
|
zinc finger protein 490 |
| chr1_+_95616933 | 0.17 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr3_-_156840776 | 0.16 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr3_-_10334617 | 0.16 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.8 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr5_+_150404904 | 0.16 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr1_-_77685084 | 0.14 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr10_+_91461337 | 0.14 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr19_-_42894420 | 0.14 |
ENST00000597255.1
ENST00000222032.5 |
CNFN
|
cornifelin |
| chr14_-_68066849 | 0.14 |
ENST00000558493.1
ENST00000561272.1 |
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr1_-_60392452 | 0.14 |
ENST00000371204.3
|
CYP2J2
|
cytochrome P450, family 2, subfamily J, polypeptide 2 |
| chr13_+_115000521 | 0.14 |
ENST00000252457.5
ENST00000375308.1 |
CDC16
|
cell division cycle 16 |
| chr1_+_156338993 | 0.14 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr16_-_3068171 | 0.13 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr19_+_54609230 | 0.13 |
ENST00000420296.1
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr19_-_44174330 | 0.13 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr1_+_186798073 | 0.13 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr20_+_2821366 | 0.12 |
ENST00000453689.1
ENST00000417508.1 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chrX_+_153237740 | 0.12 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chr15_-_79576156 | 0.12 |
ENST00000560452.1
ENST00000560872.1 ENST00000560732.1 ENST00000559979.1 ENST00000560533.1 ENST00000559225.1 |
RP11-17L5.4
|
RP11-17L5.4 |
| chr21_-_46964306 | 0.12 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.1 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr14_+_59951161 | 0.12 |
ENST00000261247.9
ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP
|
JNK1/MAPK8-associated membrane protein |
| chr16_-_66864806 | 0.12 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr5_-_1799864 | 0.12 |
ENST00000510999.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr15_+_85147127 | 0.11 |
ENST00000541040.1
ENST00000538076.1 ENST00000485222.2 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr8_+_27169138 | 0.11 |
ENST00000522338.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr14_+_97925151 | 0.11 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr5_+_34915444 | 0.11 |
ENST00000336767.5
|
BRIX1
|
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr6_-_154751629 | 0.11 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr22_+_18632666 | 0.11 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr5_-_18746250 | 0.11 |
ENST00000512978.1
|
CTD-2023M8.1
|
CTD-2023M8.1 |
| chr17_-_55911970 | 0.11 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr5_-_43412418 | 0.11 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr1_-_39339777 | 0.10 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr5_-_77072085 | 0.10 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr17_+_67498396 | 0.10 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_+_96211643 | 0.10 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr7_+_79765071 | 0.10 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr2_+_64068844 | 0.10 |
ENST00000337130.5
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr11_-_89956461 | 0.09 |
ENST00000320585.6
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr19_+_52074502 | 0.09 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr2_-_187367356 | 0.09 |
ENST00000595956.1
|
AC018867.2
|
AC018867.2 |
| chr2_+_64069016 | 0.09 |
ENST00000488245.2
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr7_+_23221438 | 0.09 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr1_-_160232312 | 0.09 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr7_+_110731062 | 0.09 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr10_-_104953009 | 0.09 |
ENST00000470299.1
ENST00000343289.5 |
NT5C2
|
5'-nucleotidase, cytosolic II |
| chrX_+_51942963 | 0.09 |
ENST00000375625.3
|
RP11-363G10.2
|
RP11-363G10.2 |
| chr20_+_2795626 | 0.09 |
ENST00000603872.1
ENST00000380589.4 |
C20orf141
|
chromosome 20 open reading frame 141 |
| chr13_-_48669232 | 0.09 |
ENST00000258648.2
ENST00000378586.1 |
MED4
|
mediator complex subunit 4 |
| chr5_+_64920826 | 0.09 |
ENST00000438419.2
ENST00000231526.4 ENST00000505553.1 ENST00000545191.1 |
TRAPPC13
|
trafficking protein particle complex 13 |
| chr7_+_142374104 | 0.09 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr8_+_22414182 | 0.09 |
ENST00000524057.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr19_+_50433476 | 0.09 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr5_+_68485363 | 0.09 |
ENST00000283006.2
ENST00000515001.1 |
CENPH
|
centromere protein H |
| chr14_+_95078714 | 0.08 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr1_-_108231101 | 0.08 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr11_-_9482010 | 0.08 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr16_+_57406368 | 0.08 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr6_+_63921399 | 0.08 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr12_-_39300071 | 0.08 |
ENST00000550863.1
|
CPNE8
|
copine VIII |
| chrX_-_38186811 | 0.08 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr6_-_83775489 | 0.08 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr19_-_45457264 | 0.08 |
ENST00000591646.1
|
CTB-129P6.11
|
Uncharacterized protein |
| chrX_-_51797351 | 0.08 |
ENST00000375644.3
|
RP11-114H20.1
|
RP11-114H20.1 |
| chr1_+_27114589 | 0.07 |
ENST00000431541.1
ENST00000449950.2 ENST00000374145.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr6_+_108616054 | 0.07 |
ENST00000368977.4
|
LACE1
|
lactation elevated 1 |
| chr13_+_115000556 | 0.07 |
ENST00000252458.6
|
CDC16
|
cell division cycle 16 |
| chr3_-_160117301 | 0.07 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr1_-_193029192 | 0.07 |
ENST00000417752.1
ENST00000367452.4 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr9_-_104145795 | 0.07 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr16_-_3074231 | 0.07 |
ENST00000572355.1
ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr3_+_124223586 | 0.07 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr17_-_39526052 | 0.07 |
ENST00000251646.3
|
KRT33B
|
keratin 33B |
| chr11_+_9482551 | 0.07 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr13_-_48669204 | 0.07 |
ENST00000417167.1
|
MED4
|
mediator complex subunit 4 |
| chr1_-_149400542 | 0.07 |
ENST00000392948.2
|
HIST2H3PS2
|
histone cluster 2, H3, pseudogene 2 |
| chr14_-_68066975 | 0.07 |
ENST00000559581.1
ENST00000560722.1 ENST00000559415.1 ENST00000216452.4 |
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr1_+_218683438 | 0.07 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr5_+_176873446 | 0.07 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr19_-_2944907 | 0.07 |
ENST00000314531.4
|
ZNF77
|
zinc finger protein 77 |
| chr14_-_74551096 | 0.07 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr2_-_162931052 | 0.06 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr6_+_26020672 | 0.06 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr11_+_94277017 | 0.06 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr4_-_77996032 | 0.06 |
ENST00000505609.1
|
CCNI
|
cyclin I |
| chr8_-_97247759 | 0.06 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr17_+_68101117 | 0.06 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_-_57087042 | 0.06 |
ENST00000317483.3
|
RAB23
|
RAB23, member RAS oncogene family |
| chr18_+_61445205 | 0.06 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr8_-_38034192 | 0.06 |
ENST00000520755.1
|
LSM1
|
LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chrX_+_71353499 | 0.06 |
ENST00000373677.1
|
NHSL2
|
NHS-like 2 |
| chr5_-_1799965 | 0.06 |
ENST00000508987.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chrX_-_153637612 | 0.06 |
ENST00000369807.1
ENST00000369808.3 |
DNASE1L1
|
deoxyribonuclease I-like 1 |
| chr5_+_75904950 | 0.06 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr7_-_143582450 | 0.06 |
ENST00000485416.1
|
FAM115A
|
family with sequence similarity 115, member A |
| chr8_-_27168737 | 0.06 |
ENST00000521253.1
ENST00000305364.4 |
TRIM35
|
tripartite motif containing 35 |
| chr20_-_271009 | 0.06 |
ENST00000382369.5
|
C20orf96
|
chromosome 20 open reading frame 96 |
| chr3_+_100428188 | 0.05 |
ENST00000418917.2
ENST00000490574.1 |
TFG
|
TRK-fused gene |
| chr10_-_32345305 | 0.05 |
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B |
| chr14_+_74551650 | 0.05 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr14_-_36536396 | 0.05 |
ENST00000546376.1
|
RP11-116N8.4
|
RP11-116N8.4 |
| chr2_+_64069459 | 0.05 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr7_+_107301447 | 0.05 |
ENST00000440056.1
|
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr6_-_135271219 | 0.05 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr1_-_203274387 | 0.05 |
ENST00000432511.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr12_-_89920030 | 0.05 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr2_-_102003987 | 0.05 |
ENST00000324768.5
|
CREG2
|
cellular repressor of E1A-stimulated genes 2 |
| chr6_-_160210692 | 0.05 |
ENST00000538128.1
ENST00000537390.1 |
TCP1
|
t-complex 1 |
| chrX_-_107681633 | 0.05 |
ENST00000394872.2
ENST00000334504.7 |
COL4A6
|
collagen, type IV, alpha 6 |
| chr11_-_76367756 | 0.05 |
ENST00000531511.1
|
RP11-672A2.4
|
RP11-672A2.4 |
| chr5_-_1799932 | 0.05 |
ENST00000382647.7
ENST00000505059.2 |
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr1_-_48866517 | 0.05 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr1_-_22109484 | 0.05 |
ENST00000529637.1
|
USP48
|
ubiquitin specific peptidase 48 |
| chr12_-_53045948 | 0.05 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr14_+_86401039 | 0.04 |
ENST00000557195.1
|
CTD-2341M24.1
|
CTD-2341M24.1 |
| chr12_+_113229337 | 0.04 |
ENST00000552667.1
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr14_-_54420133 | 0.04 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr17_-_29624343 | 0.04 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr11_-_33913708 | 0.04 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr6_+_143999072 | 0.04 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr17_-_34257731 | 0.04 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr18_+_29769978 | 0.04 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr20_+_2795609 | 0.04 |
ENST00000554164.1
ENST00000380593.4 |
TMEM239
TMEM239
|
transmembrane protein 239 CDNA FLJ26142 fis, clone TST04526; Transmembrane protein 239; Uncharacterized protein |
| chr6_-_135271260 | 0.04 |
ENST00000265605.2
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr14_-_107035208 | 0.04 |
ENST00000390626.2
|
IGHV5-51
|
immunoglobulin heavy variable 5-51 |
| chr14_+_91709103 | 0.04 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr16_+_28985542 | 0.04 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr14_-_39417410 | 0.04 |
ENST00000557019.1
ENST00000556116.1 ENST00000554732.1 |
LINC00639
|
long intergenic non-protein coding RNA 639 |
| chr7_+_7222233 | 0.04 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr20_+_2821340 | 0.04 |
ENST00000380445.3
ENST00000380469.3 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr12_+_113416340 | 0.04 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr2_-_55496476 | 0.04 |
ENST00000441307.1
|
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr9_+_134065506 | 0.04 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr7_+_141463897 | 0.04 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr1_-_110155671 | 0.04 |
ENST00000351050.3
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr3_+_156799587 | 0.04 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr4_+_159131596 | 0.04 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr1_-_216596738 | 0.04 |
ENST00000307340.3
ENST00000366943.2 ENST00000366942.3 |
USH2A
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr12_-_89919765 | 0.04 |
ENST00000541909.1
ENST00000313546.3 |
POC1B
|
POC1 centriolar protein B |
| chr7_-_7680601 | 0.04 |
ENST00000396682.2
|
RPA3
|
replication protein A3, 14kDa |
| chr17_+_41150290 | 0.04 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr1_+_59250815 | 0.04 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr15_+_44084040 | 0.04 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr1_-_154832316 | 0.03 |
ENST00000361147.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr17_+_33914424 | 0.03 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr3_+_160117418 | 0.03 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr10_+_91461413 | 0.03 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr3_-_121379739 | 0.03 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr20_-_50722183 | 0.03 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr3_+_100428268 | 0.03 |
ENST00000240851.4
|
TFG
|
TRK-fused gene |
| chr1_-_22109682 | 0.03 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr5_-_9903938 | 0.03 |
ENST00000511616.1
|
CTD-2143L24.1
|
CTD-2143L24.1 |
| chr9_+_134065519 | 0.03 |
ENST00000531600.1
|
NUP214
|
nucleoporin 214kDa |
| chr1_+_161475208 | 0.03 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr17_+_48610042 | 0.03 |
ENST00000503246.1
|
EPN3
|
epsin 3 |
| chr15_+_44084503 | 0.03 |
ENST00000409960.2
ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2
|
small EDRK-rich factor 2 |
| chr4_+_76481258 | 0.03 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chrX_-_38186775 | 0.03 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr19_-_15544099 | 0.03 |
ENST00000599910.2
|
WIZ
|
widely interspaced zinc finger motifs |
| chr19_+_16308659 | 0.03 |
ENST00000590263.1
ENST00000590756.1 ENST00000541844.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr11_+_7506713 | 0.03 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr14_-_65409438 | 0.03 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr19_-_14889349 | 0.03 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr18_-_3845321 | 0.03 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr14_-_39417473 | 0.03 |
ENST00000553932.1
|
LINC00639
|
long intergenic non-protein coding RNA 639 |
| chr7_-_6066183 | 0.03 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr3_+_93698974 | 0.03 |
ENST00000535334.1
ENST00000478400.1 ENST00000303097.7 ENST00000394222.3 ENST00000471138.1 ENST00000539730.1 |
ARL13B
|
ADP-ribosylation factor-like 13B |
| chr4_+_56719782 | 0.03 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr6_+_26124373 | 0.03 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr10_-_27529486 | 0.03 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr17_+_68100989 | 0.03 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr5_-_176889381 | 0.02 |
ENST00000393563.4
ENST00000512501.1 |
DBN1
|
drebrin 1 |
| chr10_+_32735030 | 0.02 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr3_+_52489503 | 0.02 |
ENST00000345716.4
|
NISCH
|
nischarin |
| chr19_-_44174305 | 0.02 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr4_-_4228616 | 0.02 |
ENST00000296358.4
|
OTOP1
|
otopetrin 1 |
| chr11_+_47293795 | 0.02 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chrX_+_13671225 | 0.02 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr21_-_27107198 | 0.02 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:1904344 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.2 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0051836 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |