Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
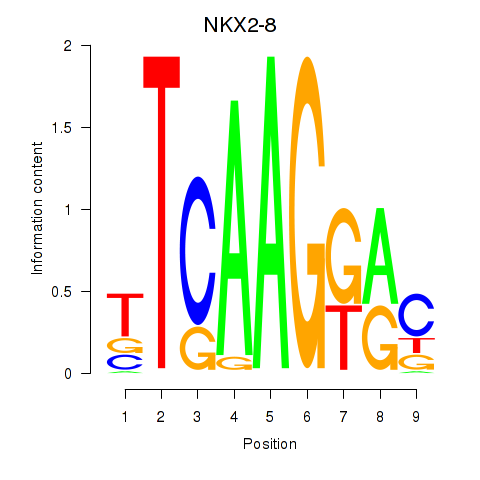
Results for NKX2-8
Z-value: 0.23
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | -0.94 | 5.5e-03 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_36779411 | 0.22 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr12_-_102591604 | 0.12 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr21_+_33671264 | 0.12 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr10_+_115674530 | 0.11 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr3_+_187930719 | 0.10 |
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr19_-_17622269 | 0.09 |
ENST00000595116.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr4_+_141264597 | 0.08 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr2_-_159313214 | 0.07 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr5_+_35852797 | 0.06 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr3_-_180397256 | 0.06 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr1_-_155006224 | 0.06 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr3_-_187455680 | 0.06 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr6_+_100054606 | 0.06 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr22_+_29834572 | 0.06 |
ENST00000354373.2
|
RFPL1
|
ret finger protein-like 1 |
| chr1_-_155006254 | 0.06 |
ENST00000295536.5
|
DCST2
|
DC-STAMP domain containing 2 |
| chr3_-_178984759 | 0.06 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr2_-_151395525 | 0.06 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr6_+_116782527 | 0.05 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr18_-_44497308 | 0.05 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr1_+_109289279 | 0.05 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr1_+_76251912 | 0.05 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr13_-_23949671 | 0.05 |
ENST00000402364.1
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr14_+_58894404 | 0.05 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr5_+_68665608 | 0.05 |
ENST00000509734.1
ENST00000354868.5 ENST00000521422.1 ENST00000354312.3 ENST00000345306.6 |
RAD17
|
RAD17 homolog (S. pombe) |
| chr5_-_179107975 | 0.05 |
ENST00000376974.4
|
CBY3
|
chibby homolog 3 (Drosophila) |
| chr1_+_120839412 | 0.05 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr4_-_100485143 | 0.05 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr14_-_58894223 | 0.05 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr12_+_16109519 | 0.04 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr14_+_21569245 | 0.04 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr14_+_37667230 | 0.04 |
ENST00000556451.1
ENST00000556753.1 ENST00000396294.2 |
MIPOL1
|
mirror-image polydactyly 1 |
| chrM_+_8527 | 0.04 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chrX_+_123094369 | 0.04 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr2_-_110371720 | 0.04 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr9_+_35909478 | 0.04 |
ENST00000443779.1
|
LINC00961
|
long intergenic non-protein coding RNA 961 |
| chr18_+_43304092 | 0.04 |
ENST00000321925.4
ENST00000587601.1 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr17_-_9808887 | 0.04 |
ENST00000226193.5
|
RCVRN
|
recoverin |
| chr7_-_99277610 | 0.04 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr8_-_81083341 | 0.04 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr3_-_196242233 | 0.04 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr9_-_99637820 | 0.03 |
ENST00000289032.8
ENST00000535338.1 |
ZNF782
|
zinc finger protein 782 |
| chr15_-_28419569 | 0.03 |
ENST00000569772.1
|
HERC2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chrX_+_123094672 | 0.03 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr7_-_137686791 | 0.03 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr14_+_37667118 | 0.03 |
ENST00000556615.1
ENST00000327441.7 ENST00000536774.1 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr8_-_29120604 | 0.03 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr7_-_5998714 | 0.03 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr16_+_58426296 | 0.03 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr11_+_71791849 | 0.03 |
ENST00000423494.2
ENST00000539587.1 ENST00000538478.1 ENST00000324866.7 ENST00000439209.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chrM_+_8366 | 0.03 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr2_-_55277692 | 0.03 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr17_+_62503073 | 0.03 |
ENST00000580188.1
ENST00000581056.1 |
CEP95
|
centrosomal protein 95kDa |
| chr3_+_187930491 | 0.03 |
ENST00000443217.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr13_+_28712614 | 0.03 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr8_-_144679264 | 0.03 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr4_+_144303093 | 0.03 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chrX_+_115567767 | 0.03 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr16_+_89696692 | 0.03 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr13_-_46425865 | 0.03 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr6_+_43968306 | 0.03 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr4_-_819901 | 0.03 |
ENST00000304062.6
|
CPLX1
|
complexin 1 |
| chr8_-_61429315 | 0.03 |
ENST00000530725.1
ENST00000532232.1 |
RP11-163N6.2
|
RP11-163N6.2 |
| chr13_+_108922228 | 0.03 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr8_+_105602961 | 0.03 |
ENST00000521923.1
|
RP11-127H5.1
|
Uncharacterized protein |
| chr3_+_39093481 | 0.03 |
ENST00000302313.5
ENST00000544962.1 ENST00000396258.3 ENST00000418020.1 |
WDR48
|
WD repeat domain 48 |
| chr17_+_73642486 | 0.03 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr3_+_239652 | 0.03 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr17_+_39994032 | 0.03 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr5_-_78808617 | 0.03 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr3_+_187930429 | 0.03 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr22_+_20877924 | 0.03 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr14_+_58765103 | 0.02 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr13_+_108921977 | 0.02 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chrX_-_69479654 | 0.02 |
ENST00000374519.2
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr12_-_21810726 | 0.02 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr12_+_57390745 | 0.02 |
ENST00000600202.1
|
HBCBP
|
HBcAg-binding protein; Uncharacterized protein |
| chr15_+_51973550 | 0.02 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr2_-_207024233 | 0.02 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr9_+_109685630 | 0.02 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr6_-_137113604 | 0.02 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr15_-_77363375 | 0.02 |
ENST00000559494.1
|
TSPAN3
|
tetraspanin 3 |
| chr14_-_24732368 | 0.02 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr12_-_21810765 | 0.02 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr17_-_28661065 | 0.02 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr1_+_47137445 | 0.02 |
ENST00000569393.1
ENST00000334122.4 ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chr8_-_125551278 | 0.02 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr1_+_16083123 | 0.02 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr14_-_58894332 | 0.02 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr18_+_13382553 | 0.02 |
ENST00000586222.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr15_-_77363441 | 0.02 |
ENST00000346495.2
ENST00000424443.3 ENST00000561277.1 |
TSPAN3
|
tetraspanin 3 |
| chr14_+_76618242 | 0.02 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr14_+_58894141 | 0.02 |
ENST00000423743.3
|
KIAA0586
|
KIAA0586 |
| chr12_+_50505762 | 0.02 |
ENST00000550487.1
ENST00000317943.2 |
COX14
|
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr12_-_46663734 | 0.02 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr22_-_30925150 | 0.02 |
ENST00000437871.1
|
SEC14L6
|
SEC14-like 6 (S. cerevisiae) |
| chr6_+_111408698 | 0.02 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr1_-_115292591 | 0.02 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr12_-_10320256 | 0.02 |
ENST00000538745.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr3_-_114343768 | 0.02 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_31830655 | 0.02 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr2_+_11752379 | 0.02 |
ENST00000396123.1
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr19_-_51893827 | 0.02 |
ENST00000574814.1
|
CTD-2616J11.4
|
chromosome 19 open reading frame 84 |
| chr20_-_43936937 | 0.02 |
ENST00000342716.4
ENST00000353917.5 ENST00000360607.6 ENST00000372751.4 |
MATN4
|
matrilin 4 |
| chr15_+_40453204 | 0.02 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr7_+_100318423 | 0.02 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr1_+_16348525 | 0.02 |
ENST00000331433.4
|
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr8_-_29120580 | 0.02 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr14_+_58765305 | 0.02 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_-_207024134 | 0.02 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr12_+_13349650 | 0.02 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr16_-_66952779 | 0.02 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr10_+_74653330 | 0.02 |
ENST00000334011.5
|
OIT3
|
oncoprotein induced transcript 3 |
| chr9_+_96051469 | 0.02 |
ENST00000453718.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr5_-_95158644 | 0.02 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr3_-_114343039 | 0.02 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr22_+_31518938 | 0.02 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr1_+_224301787 | 0.02 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr14_-_106791536 | 0.02 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr2_+_177502438 | 0.02 |
ENST00000443670.1
|
AC017048.4
|
long intergenic non-protein coding RNA 1117 |
| chr16_+_4606347 | 0.02 |
ENST00000444310.4
|
C16orf96
|
chromosome 16 open reading frame 96 |
| chr11_-_72353451 | 0.02 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr2_-_178129853 | 0.02 |
ENST00000397062.3
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr12_+_51442101 | 0.02 |
ENST00000550929.1
ENST00000262055.4 ENST00000550442.1 ENST00000549340.1 ENST00000548209.1 ENST00000548251.1 ENST00000550814.1 ENST00000547660.1 ENST00000380123.2 ENST00000548401.1 ENST00000418425.2 ENST00000547008.1 ENST00000552739.1 |
LETMD1
|
LETM1 domain containing 1 |
| chr14_-_58893876 | 0.02 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr11_-_10828892 | 0.01 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr12_-_54694758 | 0.01 |
ENST00000553070.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr14_+_58894103 | 0.01 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr2_+_136289030 | 0.01 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr2_-_3504587 | 0.01 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr15_+_93749295 | 0.01 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr2_-_152118352 | 0.01 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr11_-_10715117 | 0.01 |
ENST00000527509.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr16_+_67034466 | 0.01 |
ENST00000535696.1
|
CES4A
|
carboxylesterase 4A |
| chr12_-_95397442 | 0.01 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr14_+_37667193 | 0.01 |
ENST00000539062.2
|
MIPOL1
|
mirror-image polydactyly 1 |
| chr20_-_44485835 | 0.01 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr5_+_74011328 | 0.01 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr1_+_41204506 | 0.01 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr6_+_32146131 | 0.01 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr1_+_113010056 | 0.01 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr6_+_143772060 | 0.01 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chrX_-_64196351 | 0.01 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr11_-_133826852 | 0.01 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr7_-_95025661 | 0.01 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr14_-_78083112 | 0.01 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr11_+_27062272 | 0.01 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr4_-_100485183 | 0.01 |
ENST00000394876.2
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr10_+_24755416 | 0.01 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr4_-_70080449 | 0.01 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr14_-_106552755 | 0.01 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr7_+_30811004 | 0.01 |
ENST00000265299.6
|
FAM188B
|
family with sequence similarity 188, member B |
| chr14_+_58894706 | 0.01 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr7_+_18330035 | 0.01 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chr17_+_62503147 | 0.01 |
ENST00000553412.1
|
CEP95
|
centrosomal protein 95kDa |
| chr12_-_75603236 | 0.01 |
ENST00000540018.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr17_-_33390667 | 0.01 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr3_+_4535025 | 0.01 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr5_+_14664762 | 0.01 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chrX_-_72095622 | 0.01 |
ENST00000290273.5
|
DMRTC1
|
DMRT-like family C1 |
| chr2_-_207023918 | 0.01 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr22_-_21213676 | 0.01 |
ENST00000449120.1
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr6_-_18249971 | 0.01 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr12_-_49504623 | 0.01 |
ENST00000550137.1
ENST00000547382.1 |
LMBR1L
|
limb development membrane protein 1-like |
| chr11_-_72353494 | 0.01 |
ENST00000544570.1
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr2_-_55277436 | 0.01 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr16_+_8807419 | 0.01 |
ENST00000565016.1
ENST00000567812.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr2_+_62132781 | 0.01 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr6_+_84222194 | 0.01 |
ENST00000536636.1
|
PRSS35
|
protease, serine, 35 |
| chrX_-_64196376 | 0.01 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr15_+_45422178 | 0.01 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr11_+_59705928 | 0.01 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr20_+_58296265 | 0.01 |
ENST00000395636.2
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr8_+_125551338 | 0.01 |
ENST00000276689.3
ENST00000518008.1 ENST00000522532.1 ENST00000517367.1 |
NDUFB9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa |
| chr3_-_121740969 | 0.01 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr2_-_133429091 | 0.01 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr2_+_207024306 | 0.01 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr10_-_101673782 | 0.01 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr11_-_19223523 | 0.01 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr6_+_125524785 | 0.01 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr21_-_35899113 | 0.01 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr1_+_173793777 | 0.01 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr11_-_128457446 | 0.01 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr11_-_77348796 | 0.01 |
ENST00000263309.3
ENST00000525064.1 |
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A |
| chr6_-_32731299 | 0.01 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr8_+_128426535 | 0.01 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr17_-_46178527 | 0.01 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr12_-_75603202 | 0.01 |
ENST00000393288.2
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chrX_-_32173579 | 0.01 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr19_+_44037546 | 0.01 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr2_-_136288740 | 0.01 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr8_-_145018905 | 0.01 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr12_-_118406777 | 0.01 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr15_-_77363513 | 0.01 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr16_+_88872176 | 0.01 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr1_+_41157361 | 0.01 |
ENST00000427410.2
ENST00000447388.3 ENST00000425457.2 ENST00000453631.1 ENST00000456393.2 |
NFYC
|
nuclear transcription factor Y, gamma |
| chrX_-_9754333 | 0.01 |
ENST00000447366.1
|
GPR143
|
G protein-coupled receptor 143 |
| chr6_+_158957431 | 0.01 |
ENST00000367090.3
|
TMEM181
|
transmembrane protein 181 |
| chr19_+_32836499 | 0.01 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr18_-_72920372 | 0.01 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr1_+_55464600 | 0.01 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.0 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |