Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
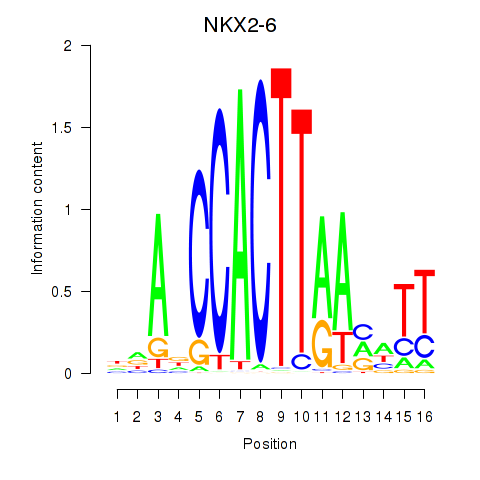
Results for NKX2-6
Z-value: 1.00
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.6 | NK2 homeobox 6 |
Activity profile of NKX2-6 motif
Sorted Z-values of NKX2-6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_187367356 | 0.99 |
ENST00000595956.1
|
AC018867.2
|
AC018867.2 |
| chr3_+_156807663 | 0.93 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr7_-_91509972 | 0.69 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr2_-_36779411 | 0.65 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr22_-_38480100 | 0.60 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr11_+_65265141 | 0.58 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr3_+_171561127 | 0.54 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr13_-_30951282 | 0.53 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr2_-_112237835 | 0.51 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr21_+_38888740 | 0.48 |
ENST00000597817.1
|
AP001421.1
|
Uncharacterized protein |
| chr19_+_52076425 | 0.47 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr19_-_43835582 | 0.42 |
ENST00000595748.1
|
CTC-490G23.2
|
CTC-490G23.2 |
| chr17_-_57229155 | 0.42 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr14_-_30766223 | 0.42 |
ENST00000549360.1
ENST00000508469.2 |
CTD-2251F13.1
|
CTD-2251F13.1 |
| chr1_-_31902614 | 0.42 |
ENST00000596131.1
|
AC114494.1
|
HCG1787699; Uncharacterized protein |
| chr11_+_59705928 | 0.34 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr7_-_37382360 | 0.34 |
ENST00000455119.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr14_+_52164675 | 0.34 |
ENST00000555936.1
|
FRMD6
|
FERM domain containing 6 |
| chr3_-_42003613 | 0.33 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr7_+_141463897 | 0.32 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr4_+_70916119 | 0.31 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr1_-_146989697 | 0.30 |
ENST00000437831.1
|
LINC00624
|
long intergenic non-protein coding RNA 624 |
| chr4_-_14889791 | 0.30 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr14_+_65182395 | 0.30 |
ENST00000554088.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr13_+_110958124 | 0.29 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr11_-_104480019 | 0.29 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr6_-_32977345 | 0.28 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr7_-_46736720 | 0.28 |
ENST00000451905.1
|
AC011294.3
|
Uncharacterized protein |
| chr1_-_17231271 | 0.26 |
ENST00000606899.1
|
RP11-108M9.6
|
RP11-108M9.6 |
| chr12_-_15103621 | 0.25 |
ENST00000536592.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr3_+_42544084 | 0.25 |
ENST00000543411.1
ENST00000438259.2 ENST00000439731.1 ENST00000325123.4 |
VIPR1
|
vasoactive intestinal peptide receptor 1 |
| chr17_-_7216939 | 0.23 |
ENST00000573684.1
|
GPS2
|
G protein pathway suppressor 2 |
| chr22_-_37882395 | 0.22 |
ENST00000416983.3
ENST00000424765.2 ENST00000356998.3 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_-_120365866 | 0.22 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr15_+_84841242 | 0.22 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr17_-_29624343 | 0.21 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr14_+_50779029 | 0.21 |
ENST00000245448.6
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr3_-_42003479 | 0.21 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr14_+_64680854 | 0.20 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr17_+_38334501 | 0.20 |
ENST00000541245.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr1_-_198906528 | 0.19 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr2_+_87769459 | 0.19 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr10_+_38717074 | 0.19 |
ENST00000423687.1
|
LINC00999
|
long intergenic non-protein coding RNA 999 |
| chr12_-_86230315 | 0.18 |
ENST00000361228.3
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr19_-_51611623 | 0.17 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr11_-_64527425 | 0.17 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr2_-_69870835 | 0.17 |
ENST00000409085.4
ENST00000406297.3 |
AAK1
|
AP2 associated kinase 1 |
| chr17_+_44370099 | 0.17 |
ENST00000496930.1
|
LRRC37A
|
leucine rich repeat containing 37A |
| chr19_-_49843539 | 0.17 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr9_-_215744 | 0.16 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr5_+_173763250 | 0.16 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr2_-_69870747 | 0.16 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr12_+_14561422 | 0.16 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_70142189 | 0.16 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr9_+_35906176 | 0.15 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr11_-_66360548 | 0.15 |
ENST00000333861.3
|
CCDC87
|
coiled-coil domain containing 87 |
| chr10_-_105437909 | 0.15 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr10_-_91403625 | 0.15 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr1_+_1243947 | 0.15 |
ENST00000379031.5
|
PUSL1
|
pseudouridylate synthase-like 1 |
| chr17_-_47723943 | 0.15 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr8_+_42396712 | 0.14 |
ENST00000518574.1
ENST00000417410.2 ENST00000414154.2 |
SMIM19
|
small integral membrane protein 19 |
| chr16_-_69373396 | 0.14 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr11_+_65627974 | 0.14 |
ENST00000525768.1
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr12_+_120031264 | 0.14 |
ENST00000426426.1
|
TMEM233
|
transmembrane protein 233 |
| chr19_-_49552363 | 0.14 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
| chr1_-_227505289 | 0.13 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr16_+_58783542 | 0.13 |
ENST00000500117.1
ENST00000565722.1 |
RP11-410D17.2
|
RP11-410D17.2 |
| chr1_-_21377383 | 0.13 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr8_+_99076750 | 0.13 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr19_+_3178736 | 0.13 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr17_-_6524159 | 0.13 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr15_-_55563072 | 0.13 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr8_+_99076509 | 0.12 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr22_+_23229960 | 0.12 |
ENST00000526893.1
ENST00000532223.2 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda-like polypeptide 5 |
| chr13_-_26452679 | 0.12 |
ENST00000596729.1
|
AL138815.2
|
Uncharacterized protein |
| chr1_+_171217622 | 0.12 |
ENST00000433267.1
ENST00000367750.3 |
FMO1
|
flavin containing monooxygenase 1 |
| chr2_+_102927962 | 0.11 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr1_-_108231101 | 0.11 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chrX_-_47341928 | 0.11 |
ENST00000313116.7
|
ZNF41
|
zinc finger protein 41 |
| chr19_-_6459746 | 0.11 |
ENST00000301454.4
ENST00000334510.5 |
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr7_-_56101826 | 0.11 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr5_-_59481406 | 0.11 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_-_123339418 | 0.11 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr18_+_39535239 | 0.11 |
ENST00000585528.1
|
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr1_-_21377447 | 0.10 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr11_-_129817356 | 0.10 |
ENST00000526082.1
|
PRDM10
|
PR domain containing 10 |
| chr15_-_41120896 | 0.10 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr10_+_32735030 | 0.10 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr4_+_113739244 | 0.10 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr7_+_100466433 | 0.10 |
ENST00000429658.1
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr12_+_32259696 | 0.10 |
ENST00000551848.1
|
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr14_-_31856397 | 0.09 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr1_+_202431859 | 0.09 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr2_-_27545921 | 0.09 |
ENST00000402310.1
ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr10_+_115939008 | 0.09 |
ENST00000369282.1
ENST00000251864.2 ENST00000369281.2 ENST00000422662.1 |
TDRD1
|
tudor domain containing 1 |
| chr2_+_203776937 | 0.09 |
ENST00000402905.3
ENST00000414490.1 ENST00000431787.1 ENST00000444724.1 ENST00000414857.1 ENST00000430899.1 ENST00000445120.1 ENST00000441569.1 ENST00000432024.1 ENST00000443740.1 ENST00000414439.1 ENST00000428585.1 ENST00000545253.1 ENST00000545262.1 ENST00000447539.1 ENST00000456821.2 ENST00000434998.1 ENST00000320443.8 |
CARF
|
calcium responsive transcription factor |
| chr8_+_20054878 | 0.09 |
ENST00000276390.2
ENST00000519667.1 |
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
| chr17_+_33914460 | 0.09 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr12_-_56236690 | 0.09 |
ENST00000322569.4
|
MMP19
|
matrix metallopeptidase 19 |
| chr19_-_12708113 | 0.09 |
ENST00000440366.1
|
ZNF490
|
zinc finger protein 490 |
| chr19_-_49552006 | 0.09 |
ENST00000391869.3
|
CGB1
|
chorionic gonadotropin, beta polypeptide 1 |
| chr14_+_52164820 | 0.09 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr12_+_56075330 | 0.09 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr9_+_96846740 | 0.09 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr15_-_65579177 | 0.09 |
ENST00000444347.2
ENST00000261888.6 |
PARP16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr19_-_56110859 | 0.09 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr7_-_150777920 | 0.08 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr1_+_159796589 | 0.08 |
ENST00000368104.4
|
SLAMF8
|
SLAM family member 8 |
| chr19_-_37663572 | 0.08 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chr17_+_33914276 | 0.08 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_-_9903938 | 0.08 |
ENST00000511616.1
|
CTD-2143L24.1
|
CTD-2143L24.1 |
| chr6_+_109169591 | 0.08 |
ENST00000368972.3
ENST00000392644.4 |
ARMC2
|
armadillo repeat containing 2 |
| chr17_-_62493131 | 0.08 |
ENST00000539111.2
|
POLG2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr1_+_205225319 | 0.08 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr9_-_70465758 | 0.08 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr14_+_50779071 | 0.08 |
ENST00000426751.2
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr6_-_15586238 | 0.08 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr2_-_121624973 | 0.08 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chr9_+_116263639 | 0.08 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr4_-_186317034 | 0.08 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr10_-_4720301 | 0.08 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr3_+_132316081 | 0.07 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr11_-_64684672 | 0.07 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr9_-_4859260 | 0.07 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr6_+_155443048 | 0.07 |
ENST00000535583.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr8_+_42396936 | 0.07 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr1_-_115259337 | 0.07 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr4_-_175041663 | 0.07 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr17_+_34890807 | 0.07 |
ENST00000429467.2
ENST00000592983.1 |
PIGW
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr19_+_42746927 | 0.07 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr8_+_136469684 | 0.07 |
ENST00000355849.5
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr19_+_52074502 | 0.06 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr2_+_70142232 | 0.06 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr3_-_113956425 | 0.06 |
ENST00000482457.2
|
ZNF80
|
zinc finger protein 80 |
| chr13_+_27998681 | 0.06 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr4_-_170924888 | 0.06 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr12_-_76377795 | 0.06 |
ENST00000552856.1
|
RP11-114H23.1
|
RP11-114H23.1 |
| chr7_-_6865826 | 0.06 |
ENST00000538180.1
|
CCZ1B
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr11_+_64685026 | 0.06 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr1_+_110162448 | 0.06 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr5_+_74907284 | 0.06 |
ENST00000601380.2
|
ANKDD1B
|
ankyrin repeat and death domain containing 1B |
| chr3_-_39196049 | 0.06 |
ENST00000514182.1
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr7_+_121513143 | 0.06 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr1_+_113009163 | 0.06 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr14_+_91709103 | 0.06 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr8_-_95220775 | 0.05 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr11_-_61687739 | 0.05 |
ENST00000531922.1
ENST00000301773.5 |
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr19_+_56111680 | 0.05 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr3_+_100428316 | 0.05 |
ENST00000479672.1
ENST00000476228.1 ENST00000463568.1 |
TFG
|
TRK-fused gene |
| chr19_+_17186577 | 0.05 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr11_-_8816375 | 0.05 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr11_+_27062860 | 0.05 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr7_+_128116783 | 0.05 |
ENST00000262432.8
ENST00000480046.1 |
METTL2B
|
methyltransferase like 2B |
| chr17_+_41150793 | 0.05 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr4_+_186317133 | 0.05 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr11_-_129817448 | 0.05 |
ENST00000304538.6
|
PRDM10
|
PR domain containing 10 |
| chr3_+_37035289 | 0.05 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr1_-_31538517 | 0.05 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr11_-_6640585 | 0.05 |
ENST00000533371.1
ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1
|
tripeptidyl peptidase I |
| chr7_-_150777949 | 0.05 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr5_+_140071011 | 0.05 |
ENST00000230771.3
ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr6_-_80247105 | 0.05 |
ENST00000369846.4
|
LCA5
|
Leber congenital amaurosis 5 |
| chr12_+_44152740 | 0.05 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr2_-_202298268 | 0.05 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr14_-_60337684 | 0.05 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr6_+_42584847 | 0.04 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr15_+_65843130 | 0.04 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr18_-_10791648 | 0.04 |
ENST00000583325.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr12_+_70219052 | 0.04 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr19_+_58144529 | 0.04 |
ENST00000347302.3
ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211
|
zinc finger protein 211 |
| chr11_+_118754475 | 0.04 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr17_+_66521936 | 0.04 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr5_+_140723601 | 0.04 |
ENST00000253812.6
|
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
| chr20_-_36661826 | 0.04 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr21_-_43735628 | 0.04 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr10_+_135204338 | 0.04 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr9_+_127023704 | 0.04 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr12_-_57039739 | 0.03 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr7_+_5938386 | 0.03 |
ENST00000537980.1
|
CCZ1
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) |
| chr11_+_27062502 | 0.03 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr19_-_37663332 | 0.03 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr11_+_65627865 | 0.03 |
ENST00000308110.4
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr2_-_100721923 | 0.03 |
ENST00000356421.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr1_+_110163202 | 0.03 |
ENST00000531203.1
ENST00000256578.3 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr10_-_27529486 | 0.03 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr2_+_109335929 | 0.03 |
ENST00000283195.6
|
RANBP2
|
RAN binding protein 2 |
| chr3_+_186435065 | 0.03 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr4_-_48082192 | 0.03 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr4_-_1685718 | 0.03 |
ENST00000472884.2
ENST00000489363.1 ENST00000308132.6 ENST00000463238.1 |
FAM53A
|
family with sequence similarity 53, member A |
| chr17_-_44896047 | 0.03 |
ENST00000225512.5
|
WNT3
|
wingless-type MMTV integration site family, member 3 |
| chr1_+_19578033 | 0.03 |
ENST00000330263.4
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae) |
| chr5_-_177210399 | 0.03 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr9_+_97766469 | 0.03 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr14_+_100531615 | 0.03 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr15_+_43425672 | 0.03 |
ENST00000260403.2
|
TMEM62
|
transmembrane protein 62 |
| chr3_-_15106747 | 0.02 |
ENST00000449354.2
ENST00000444840.2 ENST00000253686.2 |
MRPS25
|
mitochondrial ribosomal protein S25 |
| chrX_-_128782722 | 0.02 |
ENST00000427399.1
|
APLN
|
apelin |
| chr11_+_27062272 | 0.02 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr9_+_134065506 | 0.02 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr3_-_46249878 | 0.02 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |