Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
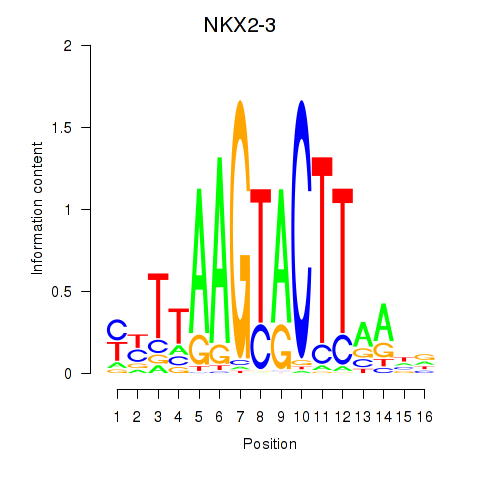
Results for NKX2-3
Z-value: 0.89
Transcription factors associated with NKX2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-3
|
ENSG00000119919.9 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-3 | hg19_v2_chr10_+_101292684_101292706 | 0.47 | 3.5e-01 | Click! |
Activity profile of NKX2-3 motif
Sorted Z-values of NKX2-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_150404904 | 1.11 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr4_-_140201333 | 0.76 |
ENST00000398955.1
|
MGARP
|
mitochondria-localized glutamic acid-rich protein |
| chr15_+_74509530 | 0.66 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr11_+_62379194 | 0.60 |
ENST00000525801.1
ENST00000534093.1 |
ROM1
|
retinal outer segment membrane protein 1 |
| chr6_+_8652370 | 0.52 |
ENST00000503668.1
|
HULC
|
hepatocellular carcinoma up-regulated long non-coding RNA |
| chr6_-_150212029 | 0.51 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr10_-_5227096 | 0.51 |
ENST00000488756.1
ENST00000334314.3 |
AKR1CL1
|
aldo-keto reductase family 1, member C-like 1 |
| chr18_-_53735601 | 0.50 |
ENST00000589754.1
|
CTD-2008L17.2
|
CTD-2008L17.2 |
| chr4_+_76439649 | 0.49 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr6_-_32160622 | 0.46 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr4_+_80584903 | 0.44 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr9_-_4859260 | 0.41 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr14_+_45366518 | 0.40 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr22_+_45714361 | 0.39 |
ENST00000452238.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr17_-_8059638 | 0.39 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr22_+_45714672 | 0.38 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr6_+_149539053 | 0.38 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr12_+_116985896 | 0.34 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr20_+_32782375 | 0.34 |
ENST00000568305.1
|
ASIP
|
agouti signaling protein |
| chrX_+_109602039 | 0.33 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr2_+_191045173 | 0.33 |
ENST00000409870.1
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chrX_-_49121165 | 0.32 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr4_-_109541539 | 0.31 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr3_+_98482175 | 0.31 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr1_-_89488510 | 0.30 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr11_+_35211511 | 0.30 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr19_+_53761545 | 0.30 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr13_-_45048386 | 0.29 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr1_-_26633480 | 0.26 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr1_+_54359854 | 0.26 |
ENST00000361921.3
ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1
|
deiodinase, iodothyronine, type I |
| chr2_+_114163945 | 0.26 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr12_-_104531945 | 0.25 |
ENST00000551446.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr1_-_240775447 | 0.25 |
ENST00000318160.4
|
GREM2
|
gremlin 2, DAN family BMP antagonist |
| chr13_-_44735393 | 0.25 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr6_+_32121789 | 0.25 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr17_-_39023462 | 0.24 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr16_+_75690093 | 0.24 |
ENST00000564671.2
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr5_-_132200477 | 0.24 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr10_+_99332529 | 0.24 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr3_+_145782358 | 0.24 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr19_-_17622684 | 0.23 |
ENST00000596643.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr10_-_97200772 | 0.23 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr5_-_110848253 | 0.23 |
ENST00000505803.1
ENST00000502322.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr5_+_140213815 | 0.23 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr3_+_149192475 | 0.23 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr10_+_99332198 | 0.22 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr22_+_45714301 | 0.22 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr6_+_131958436 | 0.22 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr7_+_7196565 | 0.22 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr4_-_39034542 | 0.22 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr12_-_42878101 | 0.21 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr22_+_25202232 | 0.21 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr6_+_32121218 | 0.21 |
ENST00000414204.1
ENST00000361568.2 ENST00000395523.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr11_+_10326612 | 0.21 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr3_-_114790179 | 0.21 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_111927062 | 0.21 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr16_-_2031464 | 0.21 |
ENST00000356120.4
ENST00000354249.4 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr11_+_33563618 | 0.21 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr1_-_26633067 | 0.21 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr13_-_46679144 | 0.21 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr19_+_50148087 | 0.20 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chrX_+_78003204 | 0.20 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr19_+_18485509 | 0.20 |
ENST00000597765.1
|
GDF15
|
growth differentiation factor 15 |
| chr12_-_52604607 | 0.20 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr18_-_60985914 | 0.19 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr16_+_31225337 | 0.19 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr19_+_56653064 | 0.18 |
ENST00000593100.1
|
ZNF444
|
zinc finger protein 444 |
| chr17_+_48823896 | 0.17 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr10_+_129785536 | 0.17 |
ENST00000419012.2
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chrX_-_115085422 | 0.17 |
ENST00000430756.2
|
RP11-761E20.1
|
RP11-761E20.1 |
| chr8_+_32579271 | 0.17 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr6_-_11382478 | 0.17 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr19_-_38743878 | 0.17 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr5_-_152069089 | 0.17 |
ENST00000506723.2
|
AC091969.1
|
AC091969.1 |
| chrX_+_71354000 | 0.17 |
ENST00000510661.1
ENST00000535692.1 |
NHSL2
|
NHS-like 2 |
| chr13_+_23993097 | 0.17 |
ENST00000443092.1
|
SACS-AS1
|
SACS antisense RNA 1 |
| chr10_-_49813090 | 0.16 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr13_-_50018241 | 0.16 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr21_+_33671160 | 0.16 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_+_113775594 | 0.16 |
ENST00000479882.1
ENST00000493014.1 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr4_-_100212132 | 0.15 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr16_-_3450963 | 0.15 |
ENST00000573327.1
ENST00000571906.1 ENST00000573830.1 ENST00000439568.2 ENST00000422427.2 ENST00000304926.3 ENST00000396852.4 |
ZSCAN32
|
zinc finger and SCAN domain containing 32 |
| chr4_+_81118647 | 0.15 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr7_-_29186008 | 0.15 |
ENST00000396276.3
ENST00000265394.5 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr3_+_124223586 | 0.15 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr9_-_117267717 | 0.15 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr19_-_15575369 | 0.15 |
ENST00000343625.7
|
RASAL3
|
RAS protein activator like 3 |
| chrX_+_71353499 | 0.15 |
ENST00000373677.1
|
NHSL2
|
NHS-like 2 |
| chr4_-_52883786 | 0.15 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr15_+_58702742 | 0.14 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr3_-_122134882 | 0.14 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr14_+_102276192 | 0.14 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_131753830 | 0.14 |
ENST00000429747.1
|
CPNE4
|
copine IV |
| chr15_-_78369994 | 0.14 |
ENST00000300584.3
ENST00000409931.3 |
TBC1D2B
|
TBC1 domain family, member 2B |
| chr3_+_190105909 | 0.14 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr11_+_94038759 | 0.14 |
ENST00000440961.2
ENST00000328458.4 |
FOLR4
|
folate receptor 4, delta (putative) |
| chr8_-_99955042 | 0.14 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr14_+_39703112 | 0.14 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr2_+_191002486 | 0.13 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr5_-_114631958 | 0.13 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr10_+_135207623 | 0.13 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr1_+_158901329 | 0.13 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr7_-_84569561 | 0.13 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr5_-_180688105 | 0.13 |
ENST00000327767.4
|
TRIM52
|
tripartite motif containing 52 |
| chr4_+_139230865 | 0.13 |
ENST00000502757.1
ENST00000507145.1 ENST00000515282.1 ENST00000510736.1 |
LINC00499
|
long intergenic non-protein coding RNA 499 |
| chr19_-_44174330 | 0.13 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr20_-_43729750 | 0.12 |
ENST00000537075.1
ENST00000306117.1 |
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr1_+_32666188 | 0.12 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chr20_+_60962143 | 0.12 |
ENST00000343986.4
|
RPS21
|
ribosomal protein S21 |
| chr15_+_85147127 | 0.12 |
ENST00000541040.1
ENST00000538076.1 ENST00000485222.2 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr12_-_104532062 | 0.12 |
ENST00000240055.3
|
NFYB
|
nuclear transcription factor Y, beta |
| chr3_-_49757101 | 0.12 |
ENST00000320431.7
|
AMIGO3
|
adhesion molecule with Ig-like domain 3 |
| chr6_-_32634425 | 0.12 |
ENST00000399082.3
ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr19_-_39368887 | 0.12 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr11_+_94277017 | 0.12 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr18_-_67624160 | 0.11 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr9_+_36036430 | 0.11 |
ENST00000377966.3
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr10_+_135207598 | 0.11 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr13_+_50018402 | 0.11 |
ENST00000354234.4
|
SETDB2
|
SET domain, bifurcated 2 |
| chr1_-_114696472 | 0.11 |
ENST00000393296.1
ENST00000369547.1 ENST00000610222.1 |
SYT6
|
synaptotagmin VI |
| chr10_-_36813162 | 0.11 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr6_-_33285505 | 0.11 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr1_+_202317855 | 0.11 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr11_+_64950801 | 0.11 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chrX_-_108976449 | 0.11 |
ENST00000469857.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr2_+_204103663 | 0.11 |
ENST00000356079.4
ENST00000429815.2 |
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr3_-_47950745 | 0.11 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr4_+_76439665 | 0.11 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr16_-_69166031 | 0.11 |
ENST00000398235.2
ENST00000520529.1 |
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr3_+_98482159 | 0.11 |
ENST00000468553.1
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr12_-_122296755 | 0.11 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr3_-_49058479 | 0.11 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr6_-_112081113 | 0.11 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr22_+_38453207 | 0.11 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr18_+_61445007 | 0.11 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr6_-_26027480 | 0.11 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr3_+_149191723 | 0.11 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr19_+_36142147 | 0.10 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr3_+_186743261 | 0.10 |
ENST00000423451.1
ENST00000446170.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr20_-_31071309 | 0.10 |
ENST00000326071.4
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr18_-_51750948 | 0.10 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr10_-_104262426 | 0.10 |
ENST00000487599.1
|
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr4_-_104020968 | 0.10 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr16_+_56969284 | 0.10 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr14_+_36295638 | 0.10 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr15_-_50558223 | 0.10 |
ENST00000267845.3
|
HDC
|
histidine decarboxylase |
| chr1_-_85870177 | 0.10 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr19_+_3178736 | 0.09 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr6_+_131148538 | 0.09 |
ENST00000541421.2
|
SMLR1
|
small leucine-rich protein 1 |
| chr12_-_131478468 | 0.09 |
ENST00000536673.1
ENST00000542980.1 |
RP11-76C10.5
|
RP11-76C10.5 |
| chr19_-_3772209 | 0.09 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr20_+_48884002 | 0.09 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr10_+_17794251 | 0.09 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr1_+_156561533 | 0.09 |
ENST00000368234.3
ENST00000368235.3 ENST00000368233.3 |
APOA1BP
|
apolipoprotein A-I binding protein |
| chr10_-_104262460 | 0.09 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr9_+_125376948 | 0.09 |
ENST00000297913.2
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1 |
| chr10_+_27793197 | 0.09 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr6_+_36562132 | 0.09 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chr7_+_1609765 | 0.09 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr17_-_80797886 | 0.08 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr17_+_12569306 | 0.08 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr17_-_36348610 | 0.08 |
ENST00000339023.4
ENST00000354664.4 |
TBC1D3
|
TBC1 domain family, member 3 |
| chr5_+_49961727 | 0.08 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr19_+_14492247 | 0.08 |
ENST00000357355.3
ENST00000592261.2 ENST00000242786.5 |
CD97
|
CD97 molecule |
| chr16_+_23765948 | 0.08 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr14_+_36295504 | 0.08 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr15_+_57891609 | 0.08 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr3_-_101039402 | 0.08 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr19_-_42746714 | 0.08 |
ENST00000222330.3
|
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr10_+_101419187 | 0.08 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr22_-_37584321 | 0.08 |
ENST00000397110.2
ENST00000337843.2 |
C1QTNF6
|
C1q and tumor necrosis factor related protein 6 |
| chr5_+_140227048 | 0.08 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr12_-_120632505 | 0.08 |
ENST00000300648.6
|
GCN1L1
|
GCN1 general control of amino-acid synthesis 1-like 1 (yeast) |
| chr6_-_33239612 | 0.08 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr13_-_46679185 | 0.08 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr1_+_230193521 | 0.08 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr18_-_64271363 | 0.08 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr16_-_66952742 | 0.08 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr3_+_30648066 | 0.08 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr20_-_33539655 | 0.08 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr11_+_118889142 | 0.07 |
ENST00000533632.1
|
TRAPPC4
|
trafficking protein particle complex 4 |
| chr7_-_105221898 | 0.07 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chr17_+_26662679 | 0.07 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr7_-_150974494 | 0.07 |
ENST00000392811.2
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr20_+_57464200 | 0.07 |
ENST00000604005.1
|
GNAS
|
GNAS complex locus |
| chr11_-_62607036 | 0.07 |
ENST00000311713.7
ENST00000278856.4 |
WDR74
|
WD repeat domain 74 |
| chr15_-_101142362 | 0.07 |
ENST00000559577.1
ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS
|
lines homolog (Drosophila) |
| chr14_+_94547675 | 0.07 |
ENST00000393115.3
ENST00000554166.1 ENST00000556381.1 ENST00000553664.1 ENST00000555341.1 ENST00000557218.1 ENST00000554544.1 ENST00000557066.1 |
IFI27L1
|
interferon, alpha-inducible protein 27-like 1 |
| chr19_-_18433910 | 0.07 |
ENST00000594828.3
ENST00000593829.1 |
LSM4
|
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr19_+_41882466 | 0.07 |
ENST00000436170.2
|
TMEM91
|
transmembrane protein 91 |
| chr9_+_118950325 | 0.07 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr18_+_43684310 | 0.07 |
ENST00000592471.1
ENST00000585518.1 |
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr11_-_64536521 | 0.07 |
ENST00000486867.1
|
SF1
|
splicing factor 1 |
| chr10_+_1120312 | 0.07 |
ENST00000436154.1
|
WDR37
|
WD repeat domain 37 |
| chr17_+_36283971 | 0.07 |
ENST00000327454.6
ENST00000378174.5 |
TBC1D3F
|
TBC1 domain family, member 3F |
| chr11_+_57435219 | 0.07 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr4_+_102734967 | 0.07 |
ENST00000444316.2
|
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr5_-_77072085 | 0.07 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr20_-_31071239 | 0.07 |
ENST00000359676.5
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr13_-_30880979 | 0.06 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.2 | 0.8 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.3 | GO:0002669 | regulation of tolerance induction dependent upon immune response(GO:0002652) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.3 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.2 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.1 | 0.5 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.2 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.2 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.6 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.1 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.5 | GO:0097503 | sialylation(GO:0097503) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 1.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.2 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.5 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |