Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
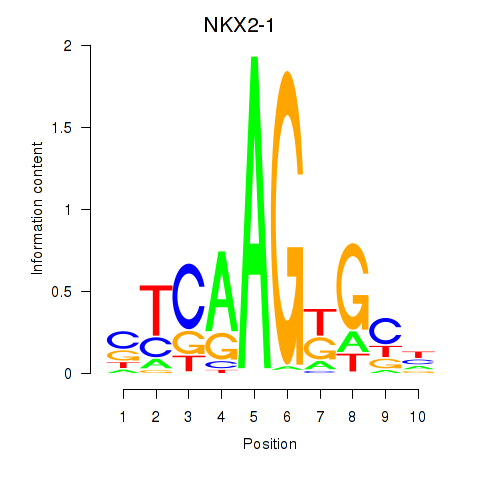
Results for NKX2-1
Z-value: 0.77
Transcription factors associated with NKX2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-1
|
ENSG00000136352.13 | NK2 homeobox 1 |
Activity profile of NKX2-1 motif
Sorted Z-values of NKX2-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39390212 | 0.50 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr20_+_825275 | 0.38 |
ENST00000541082.1
|
FAM110A
|
family with sequence similarity 110, member A |
| chr8_+_94752349 | 0.37 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr3_-_49837254 | 0.31 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr8_-_144679264 | 0.30 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr1_-_41487383 | 0.29 |
ENST00000302946.8
ENST00000372613.2 ENST00000439569.2 ENST00000397197.2 |
SLFNL1
|
schlafen-like 1 |
| chrX_-_30326445 | 0.29 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr11_+_115498761 | 0.28 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr3_+_39149145 | 0.28 |
ENST00000301819.6
ENST00000431162.2 |
TTC21A
|
tetratricopeptide repeat domain 21A |
| chr1_-_41487415 | 0.27 |
ENST00000372611.1
|
SLFNL1
|
schlafen-like 1 |
| chr11_-_86383650 | 0.26 |
ENST00000526944.1
ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr9_-_7800067 | 0.26 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chr4_+_184826418 | 0.24 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chrX_-_153236819 | 0.24 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr20_-_56285595 | 0.24 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr9_-_97356075 | 0.23 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr11_+_46193466 | 0.23 |
ENST00000533793.1
|
RP11-702F3.3
|
RP11-702F3.3 |
| chr7_-_105332084 | 0.23 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr14_+_74815116 | 0.22 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chrX_-_153236620 | 0.22 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chrX_-_153237258 | 0.22 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr19_-_36231437 | 0.21 |
ENST00000591748.1
|
IGFLR1
|
IGF-like family receptor 1 |
| chr19_-_39390350 | 0.21 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr21_+_45773515 | 0.21 |
ENST00000397932.2
ENST00000300481.9 |
TRPM2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr19_+_39390320 | 0.21 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chrX_+_22056165 | 0.21 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr21_-_36421401 | 0.20 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr2_+_85646054 | 0.20 |
ENST00000389938.2
|
SH2D6
|
SH2 domain containing 6 |
| chr1_-_153538011 | 0.20 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr1_-_17338386 | 0.19 |
ENST00000341676.5
ENST00000452699.1 |
ATP13A2
|
ATPase type 13A2 |
| chr8_-_38326119 | 0.19 |
ENST00000356207.5
ENST00000326324.6 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr21_+_33671264 | 0.19 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr17_-_3499125 | 0.18 |
ENST00000399759.3
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr5_-_146889619 | 0.18 |
ENST00000343218.5
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr18_+_13382553 | 0.18 |
ENST00000586222.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr16_+_2022036 | 0.18 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr3_+_187930719 | 0.18 |
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr18_+_42260861 | 0.18 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr19_+_48972459 | 0.18 |
ENST00000427476.1
|
CYTH2
|
cytohesin 2 |
| chr12_+_25205446 | 0.18 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr8_-_80680078 | 0.18 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr16_+_58533951 | 0.17 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr8_+_77596014 | 0.17 |
ENST00000523885.1
|
ZFHX4
|
zinc finger homeobox 4 |
| chr17_+_40610862 | 0.17 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr6_-_149806105 | 0.17 |
ENST00000389942.5
ENST00000416573.2 ENST00000542614.1 ENST00000409806.3 |
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr19_+_50148087 | 0.17 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr10_-_79397547 | 0.17 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr9_-_128246769 | 0.16 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr6_+_100054606 | 0.16 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr16_+_66638003 | 0.16 |
ENST00000562357.1
ENST00000360086.4 ENST00000562707.1 ENST00000361909.4 ENST00000460097.1 ENST00000565666.1 |
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr10_+_80027105 | 0.16 |
ENST00000461034.1
ENST00000476909.1 ENST00000459633.1 |
LINC00595
|
long intergenic non-protein coding RNA 595 |
| chr1_+_202431859 | 0.16 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr16_-_67493110 | 0.15 |
ENST00000602876.1
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr1_-_228604328 | 0.15 |
ENST00000355586.4
ENST00000366698.2 ENST00000520264.1 ENST00000479800.1 ENST00000295033.3 |
TRIM17
|
tripartite motif containing 17 |
| chr16_+_5121814 | 0.15 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr17_-_34195862 | 0.15 |
ENST00000592980.1
ENST00000587626.1 |
C17orf66
|
chromosome 17 open reading frame 66 |
| chr15_+_76030311 | 0.14 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr2_+_101591314 | 0.14 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr8_-_144679602 | 0.14 |
ENST00000526710.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrX_+_86772787 | 0.14 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr1_+_15736359 | 0.14 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chrX_+_23928500 | 0.14 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr17_+_61086917 | 0.14 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr5_-_172662303 | 0.14 |
ENST00000517440.1
ENST00000329198.4 |
NKX2-5
|
NK2 homeobox 5 |
| chr1_+_55464600 | 0.13 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr8_-_16859690 | 0.13 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr11_-_62313090 | 0.13 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr22_+_24999114 | 0.13 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr15_-_43029252 | 0.13 |
ENST00000563260.1
ENST00000356231.3 |
CDAN1
|
codanin 1 |
| chr14_+_67707826 | 0.13 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr5_-_172662230 | 0.12 |
ENST00000424406.2
|
NKX2-5
|
NK2 homeobox 5 |
| chr7_+_99156212 | 0.12 |
ENST00000454654.1
|
ZNF655
|
zinc finger protein 655 |
| chr19_+_48972265 | 0.12 |
ENST00000452733.2
|
CYTH2
|
cytohesin 2 |
| chr19_+_24009879 | 0.12 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr17_-_47492236 | 0.12 |
ENST00000434917.2
ENST00000300408.3 ENST00000511832.1 ENST00000419140.2 |
PHB
|
prohibitin |
| chr3_+_4535025 | 0.12 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr7_+_150782945 | 0.11 |
ENST00000463381.1
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr19_+_38924316 | 0.11 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chr17_+_46048376 | 0.11 |
ENST00000338399.4
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr12_+_6961279 | 0.11 |
ENST00000229268.8
ENST00000389231.5 ENST00000542087.1 |
USP5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr6_-_26056695 | 0.11 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr12_+_25205666 | 0.11 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr8_+_145203548 | 0.11 |
ENST00000534366.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr16_-_28223166 | 0.11 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr17_+_38375528 | 0.11 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr11_+_66512089 | 0.11 |
ENST00000524551.1
ENST00000525908.1 ENST00000360962.4 ENST00000346672.4 ENST00000527634.1 ENST00000540737.1 |
C11orf80
|
chromosome 11 open reading frame 80 |
| chr16_+_640055 | 0.10 |
ENST00000568586.1
ENST00000538492.1 ENST00000248139.3 |
RAB40C
|
RAB40C, member RAS oncogene family |
| chr16_-_18937072 | 0.10 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr17_-_34195889 | 0.10 |
ENST00000311880.2
|
C17orf66
|
chromosome 17 open reading frame 66 |
| chr17_-_76778339 | 0.10 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr19_+_40477062 | 0.10 |
ENST00000455878.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr2_-_225266802 | 0.10 |
ENST00000243806.2
|
FAM124B
|
family with sequence similarity 124B |
| chr17_-_49198216 | 0.10 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr17_+_46048471 | 0.10 |
ENST00000578018.1
ENST00000579175.1 |
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr20_+_30407105 | 0.10 |
ENST00000375994.2
|
MYLK2
|
myosin light chain kinase 2 |
| chr16_-_67190152 | 0.09 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr12_+_93772402 | 0.09 |
ENST00000546925.1
|
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr9_-_72374848 | 0.09 |
ENST00000377200.5
ENST00000340434.4 ENST00000472967.2 |
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr12_+_25205568 | 0.09 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr3_-_52029830 | 0.09 |
ENST00000492277.1
ENST00000475248.1 ENST00000479017.1 ENST00000495383.1 ENST00000466397.1 ENST00000481629.1 |
RPL29
|
ribosomal protein L29 |
| chr11_-_65325430 | 0.09 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr6_+_25652501 | 0.09 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr9_+_33290491 | 0.09 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr11_-_65325203 | 0.09 |
ENST00000526927.1
ENST00000536982.1 |
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr10_-_103454876 | 0.09 |
ENST00000331272.7
|
FBXW4
|
F-box and WD repeat domain containing 4 |
| chr2_-_31360887 | 0.09 |
ENST00000420311.2
ENST00000356174.3 ENST00000324589.5 |
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_-_38397384 | 0.08 |
ENST00000373027.1
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa |
| chr18_-_10791648 | 0.08 |
ENST00000583325.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr17_-_47492164 | 0.08 |
ENST00000512041.2
ENST00000446735.1 ENST00000504124.1 |
PHB
|
prohibitin |
| chr20_+_30407151 | 0.08 |
ENST00000375985.4
|
MYLK2
|
myosin light chain kinase 2 |
| chr1_+_228337553 | 0.08 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr2_-_133429091 | 0.08 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr18_+_72166564 | 0.08 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr1_-_15735925 | 0.08 |
ENST00000427824.1
|
RP3-467K16.4
|
RP3-467K16.4 |
| chr5_-_172662197 | 0.08 |
ENST00000521848.1
|
NKX2-5
|
NK2 homeobox 5 |
| chr5_+_131705438 | 0.07 |
ENST00000245407.3
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr19_-_10679644 | 0.07 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr17_-_1508379 | 0.07 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr9_-_14313641 | 0.07 |
ENST00000380953.1
|
NFIB
|
nuclear factor I/B |
| chr1_+_150266256 | 0.07 |
ENST00000309092.7
ENST00000369084.5 |
MRPS21
|
mitochondrial ribosomal protein S21 |
| chr13_-_46425865 | 0.07 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr19_+_751122 | 0.06 |
ENST00000215582.6
|
MISP
|
mitotic spindle positioning |
| chr16_+_66638567 | 0.06 |
ENST00000567572.1
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr16_+_57126428 | 0.06 |
ENST00000290776.8
|
CPNE2
|
copine II |
| chr2_+_119699864 | 0.06 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr7_+_150758642 | 0.06 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr11_+_66512303 | 0.06 |
ENST00000532565.2
|
C11orf80
|
chromosome 11 open reading frame 80 |
| chr15_+_74911430 | 0.06 |
ENST00000562670.1
ENST00000564096.1 |
CLK3
|
CDC-like kinase 3 |
| chr1_-_160232197 | 0.06 |
ENST00000419626.1
ENST00000610139.1 ENST00000475733.1 ENST00000407642.2 ENST00000368073.3 ENST00000326837.2 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr14_-_73360796 | 0.06 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr1_+_151512775 | 0.06 |
ENST00000368849.3
ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1
|
tuftelin 1 |
| chr17_+_66521936 | 0.06 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr19_+_48958766 | 0.06 |
ENST00000342291.2
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chrX_+_86772707 | 0.05 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr18_-_13915530 | 0.05 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr2_-_157189180 | 0.05 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr12_-_67197760 | 0.05 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr12_-_102591604 | 0.05 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr20_-_22565101 | 0.05 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr8_+_145065705 | 0.05 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr5_-_171615315 | 0.05 |
ENST00000176763.5
|
STK10
|
serine/threonine kinase 10 |
| chr1_-_153599426 | 0.05 |
ENST00000392622.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr19_+_36235964 | 0.05 |
ENST00000587708.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr3_-_139195350 | 0.05 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr9_+_133986782 | 0.05 |
ENST00000372301.2
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr16_+_640201 | 0.05 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr1_-_202679535 | 0.05 |
ENST00000367268.4
|
SYT2
|
synaptotagmin II |
| chr2_-_230787879 | 0.05 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr16_+_57126482 | 0.05 |
ENST00000537605.1
ENST00000535318.2 |
CPNE2
|
copine II |
| chr1_+_180165672 | 0.04 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr12_+_60058458 | 0.04 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr8_-_144679296 | 0.04 |
ENST00000317198.6
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr2_-_225266711 | 0.04 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr1_-_25291475 | 0.04 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr5_+_140430979 | 0.04 |
ENST00000306549.3
|
PCDHB1
|
protocadherin beta 1 |
| chr17_+_46048497 | 0.04 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr17_+_4613918 | 0.04 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr16_-_28222797 | 0.04 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chrX_-_70473957 | 0.04 |
ENST00000373984.3
ENST00000314425.5 ENST00000373982.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr16_+_12995614 | 0.04 |
ENST00000423335.2
|
SHISA9
|
shisa family member 9 |
| chr14_-_74892805 | 0.04 |
ENST00000331628.3
ENST00000554953.1 |
SYNDIG1L
|
synapse differentiation inducing 1-like |
| chr12_+_13349711 | 0.04 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr8_+_145065521 | 0.04 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr16_-_58663720 | 0.04 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr15_+_77287426 | 0.04 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr1_-_92952433 | 0.04 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr22_+_24891210 | 0.03 |
ENST00000382760.2
|
UPB1
|
ureidopropionase, beta |
| chr8_+_37654693 | 0.03 |
ENST00000412232.2
|
GPR124
|
G protein-coupled receptor 124 |
| chr10_+_102222798 | 0.03 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr2_+_119699742 | 0.03 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr8_-_67341208 | 0.03 |
ENST00000499642.1
|
RP11-346I3.4
|
RP11-346I3.4 |
| chr12_-_10978957 | 0.03 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr18_+_56806701 | 0.03 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr4_+_144354644 | 0.03 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr6_-_75828774 | 0.03 |
ENST00000493109.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr8_-_114389353 | 0.03 |
ENST00000343508.3
|
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr1_+_214163033 | 0.03 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr3_+_184056614 | 0.03 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr1_+_32930647 | 0.03 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr6_-_71012773 | 0.03 |
ENST00000370496.3
ENST00000357250.6 |
COL9A1
|
collagen, type IX, alpha 1 |
| chr17_-_42466864 | 0.03 |
ENST00000353281.4
ENST00000262407.5 |
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chrX_-_70474377 | 0.03 |
ENST00000373978.1
ENST00000373981.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr7_-_27224842 | 0.02 |
ENST00000517402.1
|
HOXA11
|
homeobox A11 |
| chr1_-_182369751 | 0.02 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr3_+_187930429 | 0.02 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr8_-_28747424 | 0.02 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr15_-_33360085 | 0.02 |
ENST00000334528.9
|
FMN1
|
formin 1 |
| chr3_-_114790179 | 0.02 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_128848740 | 0.02 |
ENST00000375990.3
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr16_+_66638616 | 0.02 |
ENST00000564060.1
ENST00000565922.1 |
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr1_-_175712829 | 0.02 |
ENST00000367674.2
|
TNR
|
tenascin R |
| chr15_-_94614049 | 0.02 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr10_+_82300575 | 0.02 |
ENST00000313455.4
|
SH2D4B
|
SH2 domain containing 4B |
| chr7_-_27224795 | 0.02 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chrX_-_110655306 | 0.01 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr21_+_45432174 | 0.01 |
ENST00000380221.3
ENST00000291574.4 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr16_+_1359138 | 0.01 |
ENST00000325437.5
|
UBE2I
|
ubiquitin-conjugating enzyme E2I |
| chr16_+_28303804 | 0.01 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr20_-_45142154 | 0.01 |
ENST00000347606.4
ENST00000457685.2 |
ZNF334
|
zinc finger protein 334 |
| chr10_-_129691195 | 0.01 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.2 | 0.7 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.3 | GO:0060927 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.1 | 0.2 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.2 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.1 | 0.2 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.2 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.2 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.3 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.2 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.0 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.3 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.0 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |