Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
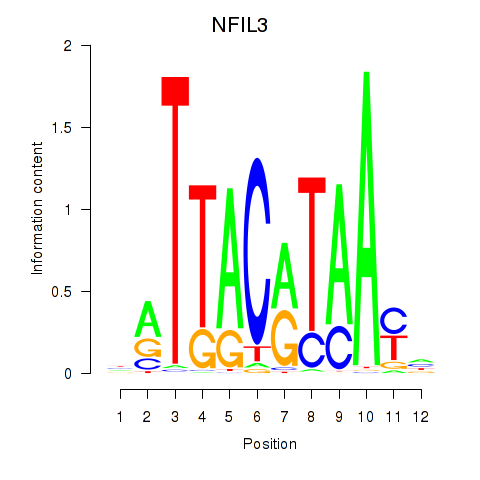
Results for NFIL3
Z-value: 0.56
Transcription factors associated with NFIL3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFIL3
|
ENSG00000165030.3 | nuclear factor, interleukin 3 regulated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIL3 | hg19_v2_chr9_-_94186131_94186174 | -0.35 | 4.9e-01 | Click! |
Activity profile of NFIL3 motif
Sorted Z-values of NFIL3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_61151306 | 0.71 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr8_+_19536083 | 0.53 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr7_+_30589829 | 0.33 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr7_+_149416439 | 0.27 |
ENST00000497895.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr14_-_62217779 | 0.26 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr20_+_34020827 | 0.25 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr18_-_53735601 | 0.25 |
ENST00000589754.1
|
CTD-2008L17.2
|
CTD-2008L17.2 |
| chr8_+_128427857 | 0.25 |
ENST00000391675.1
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr13_+_111972980 | 0.24 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr17_-_8059638 | 0.24 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr5_-_138739739 | 0.23 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr8_+_40018977 | 0.23 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr3_+_157154578 | 0.22 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr8_+_86999516 | 0.21 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr14_+_96722152 | 0.21 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr22_-_36850991 | 0.21 |
ENST00000442579.1
|
RP5-1119A7.14
|
RP5-1119A7.14 |
| chr18_+_68002675 | 0.21 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr8_-_30013748 | 0.20 |
ENST00000607315.1
|
RP11-51J9.5
|
RP11-51J9.5 |
| chr7_-_65113280 | 0.20 |
ENST00000593865.1
|
AC104057.1
|
Protein LOC100996407 |
| chr11_+_29181503 | 0.20 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr3_-_156840776 | 0.19 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr19_+_3762703 | 0.18 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr19_-_4717835 | 0.17 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr13_-_52378281 | 0.17 |
ENST00000218981.1
|
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr12_-_91573132 | 0.17 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr5_-_157161727 | 0.17 |
ENST00000599823.1
|
AC026407.1
|
Uncharacterized protein |
| chr19_-_50979981 | 0.16 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr11_-_9482010 | 0.16 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr6_+_168434678 | 0.16 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr1_-_15850676 | 0.16 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr6_+_24126350 | 0.16 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr19_-_50836762 | 0.15 |
ENST00000474951.1
ENST00000391818.2 |
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
| chr3_+_52279737 | 0.15 |
ENST00000457351.2
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr10_+_91152303 | 0.15 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr6_-_133055896 | 0.15 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr16_+_19078960 | 0.15 |
ENST00000568985.1
ENST00000566110.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr1_-_149832704 | 0.15 |
ENST00000392933.1
ENST00000369157.2 ENST00000392932.4 |
HIST2H4B
|
histone cluster 2, H4b |
| chr5_+_150404904 | 0.15 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr4_-_109541539 | 0.15 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr3_-_12587055 | 0.15 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr3_+_186330712 | 0.14 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr4_-_69536346 | 0.14 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr17_+_67590125 | 0.14 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr5_-_99870890 | 0.14 |
ENST00000499025.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr7_+_90032821 | 0.14 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr21_-_44582378 | 0.14 |
ENST00000433840.1
|
AP001631.10
|
AP001631.10 |
| chr12_+_9066472 | 0.14 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr12_-_10007448 | 0.14 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr6_+_27782788 | 0.14 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr6_+_26020672 | 0.13 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr21_+_45287112 | 0.13 |
ENST00000448287.1
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr1_-_238649319 | 0.13 |
ENST00000400946.2
|
RP11-371I1.2
|
long intergenic non-protein coding RNA 1139 |
| chr22_+_50981079 | 0.13 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr14_+_102276192 | 0.13 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr7_-_14026063 | 0.13 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr8_+_33613265 | 0.13 |
ENST00000517292.1
|
RP11-317N12.1
|
RP11-317N12.1 |
| chr21_-_40817645 | 0.13 |
ENST00000438404.1
ENST00000358268.2 ENST00000411566.1 ENST00000451131.1 ENST00000418018.1 ENST00000415863.1 ENST00000426783.1 ENST00000288350.3 ENST00000485895.2 ENST00000448288.2 ENST00000456017.1 ENST00000434281.1 |
LCA5L
|
Leber congenital amaurosis 5-like |
| chr12_-_122107549 | 0.13 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr1_-_152552980 | 0.13 |
ENST00000368787.3
|
LCE3D
|
late cornified envelope 3D |
| chr6_-_26199499 | 0.13 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr11_+_18287721 | 0.12 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr7_-_45026159 | 0.12 |
ENST00000584327.1
ENST00000438705.3 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr4_+_185734773 | 0.12 |
ENST00000508020.1
|
RP11-701P16.2
|
Uncharacterized protein |
| chr1_+_154909803 | 0.12 |
ENST00000604546.1
|
RP11-307C12.13
|
RP11-307C12.13 |
| chr5_-_149792295 | 0.12 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr1_+_149804218 | 0.12 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr5_-_176433565 | 0.12 |
ENST00000428382.2
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr1_+_228645796 | 0.12 |
ENST00000369160.2
|
HIST3H2BB
|
histone cluster 3, H2bb |
| chr15_-_49170219 | 0.12 |
ENST00000558220.1
ENST00000537958.1 |
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr19_-_1155118 | 0.12 |
ENST00000590998.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr6_-_41888843 | 0.12 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr1_-_193075180 | 0.12 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr4_+_69962185 | 0.12 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr6_-_116575226 | 0.12 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr19_-_17559376 | 0.12 |
ENST00000341130.5
|
TMEM221
|
transmembrane protein 221 |
| chr17_+_73455788 | 0.12 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr6_+_126277842 | 0.12 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr13_-_52378231 | 0.12 |
ENST00000280056.2
ENST00000444610.2 |
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr6_+_75994755 | 0.11 |
ENST00000607799.1
|
RP1-234P15.4
|
RP1-234P15.4 |
| chr10_-_7661623 | 0.11 |
ENST00000298441.6
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr7_-_45026419 | 0.11 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr7_-_45026200 | 0.11 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr19_-_7698599 | 0.11 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr15_+_89921280 | 0.11 |
ENST00000560596.1
ENST00000558692.1 ENST00000538734.2 ENST00000559235.1 |
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr10_+_6779326 | 0.11 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr11_-_104972158 | 0.11 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr16_-_4588762 | 0.11 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr4_+_80584903 | 0.11 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr4_+_70894130 | 0.11 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr2_+_162101247 | 0.11 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr3_-_49967292 | 0.11 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr19_+_38307999 | 0.10 |
ENST00000589653.1
ENST00000590433.1 |
CTD-2554C21.2
|
CTD-2554C21.2 |
| chr14_+_65453432 | 0.10 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr14_+_56584414 | 0.10 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr11_+_33563618 | 0.10 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr4_+_70146217 | 0.10 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr15_-_54267147 | 0.10 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr5_-_99870932 | 0.10 |
ENST00000504833.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chrX_+_43036243 | 0.10 |
ENST00000440955.1
|
RP3-326I13.1
|
RP3-326I13.1 |
| chr2_+_145425573 | 0.10 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr17_-_4090402 | 0.10 |
ENST00000574736.1
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr6_-_154751629 | 0.10 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr2_+_65663812 | 0.10 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr21_+_35736302 | 0.10 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr21_+_27011584 | 0.10 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr12_+_21207503 | 0.09 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr4_-_88450372 | 0.09 |
ENST00000543631.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr3_-_100565249 | 0.09 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr22_+_24115000 | 0.09 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chr1_-_26633480 | 0.09 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr6_+_26204825 | 0.09 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr19_+_36024310 | 0.09 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr8_-_83589388 | 0.09 |
ENST00000522776.1
|
RP11-653B10.1
|
RP11-653B10.1 |
| chr4_+_153021899 | 0.09 |
ENST00000509332.1
ENST00000504144.1 ENST00000499452.2 |
RP11-18H21.1
|
RP11-18H21.1 |
| chrX_+_103217207 | 0.09 |
ENST00000563257.1
ENST00000540220.1 ENST00000436583.1 ENST00000567181.1 ENST00000569577.1 |
TMSB15B
|
thymosin beta 15B |
| chr7_+_116451100 | 0.09 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr18_+_61445205 | 0.09 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr20_+_33146510 | 0.09 |
ENST00000397709.1
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr1_+_207818460 | 0.09 |
ENST00000508064.2
|
CR1L
|
complement component (3b/4b) receptor 1-like |
| chr6_-_133055815 | 0.09 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr1_-_161208013 | 0.09 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr19_-_40919271 | 0.09 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr7_-_45956856 | 0.09 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr11_-_72145426 | 0.09 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr13_+_113699029 | 0.09 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr12_-_6982442 | 0.09 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr14_+_63671105 | 0.09 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr6_+_76330355 | 0.09 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr5_-_76383133 | 0.09 |
ENST00000255198.2
|
ZBED3
|
zinc finger, BED-type containing 3 |
| chr10_-_52645416 | 0.09 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr2_+_10223908 | 0.09 |
ENST00000425235.1
|
AC104794.4
|
AC104794.4 |
| chr11_+_58938903 | 0.09 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr17_+_67498396 | 0.09 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr13_-_45048386 | 0.09 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr14_-_90097910 | 0.09 |
ENST00000550332.2
|
RP11-944C7.1
|
Protein LOC100506792 |
| chr14_+_67708344 | 0.09 |
ENST00000557237.1
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr7_+_37723336 | 0.09 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr2_+_85921414 | 0.09 |
ENST00000263863.4
ENST00000524600.1 |
GNLY
|
granulysin |
| chr2_-_46769694 | 0.09 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr1_+_222910625 | 0.09 |
ENST00000360827.2
|
FAM177B
|
family with sequence similarity 177, member B |
| chr19_+_50979753 | 0.09 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr14_-_38028689 | 0.09 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr22_-_36635598 | 0.09 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr19_-_58204128 | 0.08 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr20_+_4667094 | 0.08 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr11_-_105948040 | 0.08 |
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr16_+_77233294 | 0.08 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr11_-_117166201 | 0.08 |
ENST00000510915.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr10_-_97321112 | 0.08 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr1_-_95285652 | 0.08 |
ENST00000442418.1
|
LINC01057
|
long intergenic non-protein coding RNA 1057 |
| chr3_+_172034218 | 0.08 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr19_-_5567842 | 0.08 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr4_+_40194609 | 0.08 |
ENST00000508513.1
|
RHOH
|
ras homolog family member H |
| chr20_+_44462481 | 0.08 |
ENST00000491381.1
ENST00000342644.5 ENST00000372542.1 |
SNX21
|
sorting nexin family member 21 |
| chr1_-_193074504 | 0.08 |
ENST00000367439.3
|
GLRX2
|
glutaredoxin 2 |
| chr18_+_42277095 | 0.08 |
ENST00000591940.1
|
SETBP1
|
SET binding protein 1 |
| chr1_+_1407133 | 0.08 |
ENST00000378741.3
ENST00000308647.7 |
ATAD3B
|
ATPase family, AAA domain containing 3B |
| chr20_-_43743790 | 0.08 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr6_+_131958436 | 0.08 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr15_-_100258029 | 0.08 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr18_+_52385068 | 0.08 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr17_+_74729060 | 0.08 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr6_+_27833034 | 0.08 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr19_-_2944907 | 0.08 |
ENST00000314531.4
|
ZNF77
|
zinc finger protein 77 |
| chr14_+_93389425 | 0.08 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr16_+_532503 | 0.08 |
ENST00000412256.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr17_+_19314432 | 0.08 |
ENST00000575165.2
|
RNF112
|
ring finger protein 112 |
| chr17_-_74547256 | 0.08 |
ENST00000589145.1
|
CYGB
|
cytoglobin |
| chr3_-_128690173 | 0.08 |
ENST00000508239.1
|
RP11-723O4.6
|
Uncharacterized protein FLJ43738 |
| chr3_+_158288942 | 0.08 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr2_+_101591314 | 0.08 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr2_+_228678550 | 0.08 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr8_+_9009296 | 0.08 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr14_+_29241910 | 0.08 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr10_-_95360983 | 0.07 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr11_+_94300474 | 0.07 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr1_-_119682812 | 0.07 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr19_-_55574538 | 0.07 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr1_+_159272111 | 0.07 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr6_+_26045603 | 0.07 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr14_-_50053081 | 0.07 |
ENST00000396020.3
ENST00000245458.6 |
RPS29
|
ribosomal protein S29 |
| chr16_-_30102547 | 0.07 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr13_+_74805561 | 0.07 |
ENST00000419499.1
|
LINC00402
|
long intergenic non-protein coding RNA 402 |
| chr6_-_31940065 | 0.07 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr12_+_90313394 | 0.07 |
ENST00000549551.1
|
RP11-654D12.2
|
RP11-654D12.2 |
| chr8_-_10697281 | 0.07 |
ENST00000524114.1
ENST00000553390.1 ENST00000554914.1 |
PINX1
SOX7
SOX7
|
PIN2/TERF1 interacting, telomerase inhibitor 1 SRY (sex determining region Y)-box 7 Transcription factor SOX-7; Uncharacterized protein; cDNA FLJ58508, highly similar to Transcription factor SOX-7 |
| chr14_-_50506589 | 0.07 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr3_+_145782358 | 0.07 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr14_-_22005343 | 0.07 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr5_+_179135246 | 0.07 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr9_+_116111794 | 0.07 |
ENST00000374183.4
|
BSPRY
|
B-box and SPRY domain containing |
| chr5_-_43397184 | 0.07 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr11_-_95523500 | 0.07 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr19_+_38308119 | 0.07 |
ENST00000592103.1
|
CTD-2554C21.2
|
CTD-2554C21.2 |
| chr16_-_54962704 | 0.07 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr7_+_1126437 | 0.07 |
ENST00000413368.1
ENST00000397092.1 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr10_-_99771079 | 0.07 |
ENST00000309155.3
|
CRTAC1
|
cartilage acidic protein 1 |
| chr7_+_23719749 | 0.07 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr4_+_69962212 | 0.07 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr6_+_150690133 | 0.07 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFIL3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.0 | 0.1 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:1904744 | positive regulation of telomeric DNA binding(GO:1904744) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.0 | GO:0015920 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.0 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.0 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.3 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.0 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.0 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |