Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
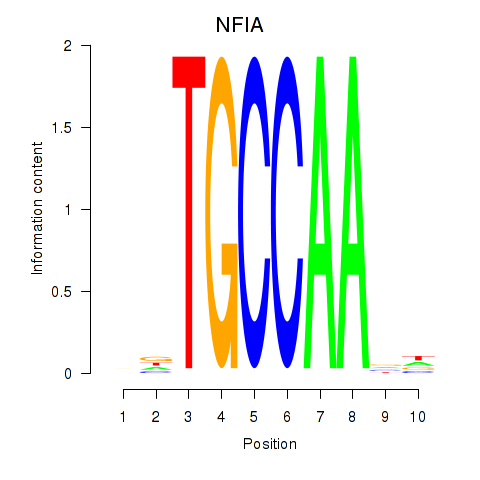
Results for NFIA
Z-value: 0.84
Transcription factors associated with NFIA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFIA
|
ENSG00000162599.11 | nuclear factor I A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIA | hg19_v2_chr1_+_61547405_61547534 | 0.94 | 5.2e-03 | Click! |
Activity profile of NFIA motif
Sorted Z-values of NFIA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_77582905 | 1.17 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr1_+_95975672 | 0.67 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr11_-_104480019 | 0.62 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr14_-_98444386 | 0.61 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr1_-_114429997 | 0.56 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr17_+_14277419 | 0.55 |
ENST00000436469.1
|
AC022816.2
|
AC022816.2 |
| chr4_+_146402346 | 0.37 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr16_+_22524379 | 0.36 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr3_+_69812877 | 0.36 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr3_-_52002403 | 0.35 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr19_+_42254885 | 0.34 |
ENST00000595740.1
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr10_-_99030395 | 0.34 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr2_+_135596180 | 0.32 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_-_8302298 | 0.32 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr3_-_131736593 | 0.31 |
ENST00000514999.1
|
CPNE4
|
copine IV |
| chr10_+_5238793 | 0.30 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr6_-_56492816 | 0.29 |
ENST00000522360.1
|
DST
|
dystonin |
| chr10_-_6104253 | 0.29 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chr17_-_74533734 | 0.29 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr16_+_21312170 | 0.28 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr6_-_30043539 | 0.27 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr17_+_19314505 | 0.27 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr1_-_114430169 | 0.27 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr3_-_33260283 | 0.25 |
ENST00000412539.1
|
SUSD5
|
sushi domain containing 5 |
| chr8_-_121824374 | 0.25 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr11_-_93271058 | 0.25 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr7_-_41742697 | 0.24 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr7_-_27135591 | 0.23 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr2_-_65659762 | 0.23 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_-_56160625 | 0.22 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr1_+_41447609 | 0.22 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chr2_+_159651821 | 0.22 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr11_+_28724129 | 0.22 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr11_-_111649015 | 0.22 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_-_31637560 | 0.22 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr2_-_158345462 | 0.21 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chrX_-_131353461 | 0.21 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chrX_+_122993544 | 0.21 |
ENST00000422098.1
|
XIAP
|
X-linked inhibitor of apoptosis |
| chr21_+_35553045 | 0.21 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr4_-_176733897 | 0.21 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr1_-_205912577 | 0.20 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr4_-_157892055 | 0.20 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr12_-_82752159 | 0.20 |
ENST00000552377.1
|
CCDC59
|
coiled-coil domain containing 59 |
| chr11_+_126262027 | 0.20 |
ENST00000526311.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr17_-_39538550 | 0.20 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr11_+_64004888 | 0.20 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr16_-_19725899 | 0.19 |
ENST00000567367.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr9_+_35806082 | 0.19 |
ENST00000447210.1
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr13_-_46742630 | 0.19 |
ENST00000416500.1
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_-_95510743 | 0.18 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr11_-_61101247 | 0.18 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chr4_+_156824840 | 0.18 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr3_-_25824353 | 0.18 |
ENST00000427041.1
|
NGLY1
|
N-glycanase 1 |
| chr16_+_14802801 | 0.18 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr6_-_32784687 | 0.18 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr6_+_106959718 | 0.17 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr6_+_108977520 | 0.17 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr11_+_111385497 | 0.17 |
ENST00000375618.4
ENST00000529167.1 ENST00000332814.6 |
C11orf88
|
chromosome 11 open reading frame 88 |
| chr9_-_127710292 | 0.17 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr2_-_231989808 | 0.17 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr11_-_122931881 | 0.17 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr2_+_33701707 | 0.17 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_-_19626351 | 0.16 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr6_+_26156551 | 0.16 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr3_+_69812701 | 0.16 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr5_-_168006324 | 0.16 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr12_+_52203789 | 0.16 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr3_+_190105909 | 0.16 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr22_+_37959647 | 0.16 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr12_+_100897130 | 0.15 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr20_+_42574317 | 0.15 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr8_+_124194752 | 0.15 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr6_+_126221034 | 0.15 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr15_-_79576156 | 0.15 |
ENST00000560452.1
ENST00000560872.1 ENST00000560732.1 ENST00000559979.1 ENST00000560533.1 ENST00000559225.1 |
RP11-17L5.4
|
RP11-17L5.4 |
| chr1_+_150980889 | 0.15 |
ENST00000450884.1
ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE
|
prune exopolyphosphatase |
| chr3_-_105588231 | 0.15 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chrX_+_122993657 | 0.15 |
ENST00000434753.3
ENST00000430625.1 |
XIAP
|
X-linked inhibitor of apoptosis |
| chr14_-_54418598 | 0.15 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr17_+_9745786 | 0.15 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr7_-_27183263 | 0.15 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr16_+_22518495 | 0.14 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr5_-_142784101 | 0.14 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr2_+_234590556 | 0.14 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr1_+_153746683 | 0.14 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr8_+_66619277 | 0.14 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chrX_+_135251835 | 0.14 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr19_-_40724246 | 0.14 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr4_+_144354644 | 0.14 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr20_-_14318248 | 0.14 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr16_-_10652993 | 0.14 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr8_+_40010989 | 0.14 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr10_+_95848824 | 0.14 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr3_+_142342240 | 0.14 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr9_+_130911770 | 0.13 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chrX_+_135252050 | 0.13 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr9_+_130911723 | 0.13 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr11_-_10822029 | 0.13 |
ENST00000528839.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr5_+_40909354 | 0.13 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr3_+_111260980 | 0.13 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr3_+_186501979 | 0.13 |
ENST00000498746.1
|
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr12_+_7169887 | 0.13 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr22_-_38539487 | 0.13 |
ENST00000498338.1
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr13_-_46716969 | 0.13 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr16_+_67840986 | 0.13 |
ENST00000561639.1
ENST00000567852.1 ENST00000565148.1 ENST00000388833.3 ENST00000561654.1 ENST00000431934.2 |
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr3_-_176914238 | 0.13 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr19_-_12997995 | 0.13 |
ENST00000264834.4
|
KLF1
|
Kruppel-like factor 1 (erythroid) |
| chr3_+_109128961 | 0.13 |
ENST00000489670.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr5_+_112074029 | 0.12 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr16_-_79634595 | 0.12 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr19_+_36249057 | 0.12 |
ENST00000301165.5
ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr2_-_99485825 | 0.12 |
ENST00000423771.1
|
KIAA1211L
|
KIAA1211-like |
| chr22_-_23922410 | 0.12 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr5_-_142784888 | 0.12 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr5_-_159766528 | 0.12 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr15_+_59499031 | 0.12 |
ENST00000307144.4
|
LDHAL6B
|
lactate dehydrogenase A-like 6B |
| chr8_+_9009296 | 0.12 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr16_-_15474904 | 0.12 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr1_+_173684047 | 0.12 |
ENST00000546011.1
ENST00000209884.4 |
KLHL20
|
kelch-like family member 20 |
| chr8_-_16859690 | 0.12 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr3_-_176914191 | 0.12 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr13_+_100634004 | 0.12 |
ENST00000376335.3
|
ZIC2
|
Zic family member 2 |
| chr8_+_1919481 | 0.12 |
ENST00000518539.2
|
KBTBD11-OT1
|
KBTBD11 overlapping transcript 1 (non-protein coding) |
| chr6_+_99282570 | 0.12 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chrX_-_106146547 | 0.12 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr5_+_89770696 | 0.12 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr14_+_70234642 | 0.11 |
ENST00000555349.1
ENST00000554021.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr5_-_142783694 | 0.11 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_+_8995832 | 0.11 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr9_+_123906331 | 0.11 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr17_+_1733251 | 0.11 |
ENST00000570451.1
|
RPA1
|
replication protein A1, 70kDa |
| chr4_-_48271802 | 0.11 |
ENST00000381501.3
|
TEC
|
tec protein tyrosine kinase |
| chr2_-_136288740 | 0.11 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr8_-_22089533 | 0.11 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr2_-_40739501 | 0.11 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr10_+_121578211 | 0.11 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr6_+_90192974 | 0.11 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr12_-_125002827 | 0.11 |
ENST00000420698.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr11_-_85430088 | 0.11 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr16_-_89119355 | 0.11 |
ENST00000537498.1
|
CTD-2555A7.2
|
CTD-2555A7.2 |
| chr5_+_148737562 | 0.11 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr2_-_191878162 | 0.11 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr17_+_27369918 | 0.11 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr8_+_9183618 | 0.11 |
ENST00000518619.1
|
RP11-115J16.1
|
RP11-115J16.1 |
| chr3_-_121740969 | 0.11 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chrX_-_13835147 | 0.10 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr3_-_171177852 | 0.10 |
ENST00000284483.8
ENST00000475336.1 ENST00000357327.5 ENST00000460047.1 ENST00000488470.1 ENST00000470834.1 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr2_-_191878681 | 0.10 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr1_+_66797687 | 0.10 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr2_-_200320768 | 0.10 |
ENST00000428695.1
|
SATB2
|
SATB homeobox 2 |
| chr3_+_69928256 | 0.10 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr15_-_52030293 | 0.10 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr17_-_74533963 | 0.10 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr11_+_118230287 | 0.10 |
ENST00000252108.3
ENST00000431736.2 |
UBE4A
|
ubiquitination factor E4A |
| chr12_+_19389814 | 0.10 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr18_+_29171689 | 0.10 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr10_+_22614547 | 0.10 |
ENST00000416820.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr13_+_97874574 | 0.10 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr14_-_64194745 | 0.10 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr5_-_55777586 | 0.10 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr2_-_74730087 | 0.10 |
ENST00000341396.2
|
LBX2
|
ladybird homeobox 2 |
| chr1_+_11839874 | 0.10 |
ENST00000449278.1
|
C1orf167
|
chromosome 1 open reading frame 167 |
| chr12_-_21910775 | 0.09 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr1_-_235098935 | 0.09 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr7_-_24957699 | 0.09 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr3_+_121613265 | 0.09 |
ENST00000295605.2
|
SLC15A2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr11_+_62556596 | 0.09 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr12_-_11150474 | 0.09 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr3_+_4535155 | 0.09 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr5_-_43313269 | 0.09 |
ENST00000511774.1
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr17_+_7462031 | 0.09 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr4_+_146402997 | 0.09 |
ENST00000512019.1
|
SMAD1
|
SMAD family member 1 |
| chr3_+_4535025 | 0.09 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr11_-_61348292 | 0.09 |
ENST00000539008.1
ENST00000540677.1 ENST00000542836.1 ENST00000542670.1 ENST00000535826.1 ENST00000545053.1 |
SYT7
|
synaptotagmin VII |
| chr8_+_9953214 | 0.09 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr19_+_13988061 | 0.09 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chr19_-_40324255 | 0.09 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr5_+_150157860 | 0.09 |
ENST00000600109.1
|
AC010441.1
|
AC010441.1 |
| chr3_+_150264555 | 0.09 |
ENST00000406576.3
ENST00000482093.1 ENST00000273435.5 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr17_-_62308087 | 0.09 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr11_-_113577052 | 0.09 |
ENST00000540540.1
ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr1_-_155224699 | 0.09 |
ENST00000491082.1
|
FAM189B
|
family with sequence similarity 189, member B |
| chr2_+_62132781 | 0.09 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr19_+_17865011 | 0.09 |
ENST00000596462.1
ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1
|
FCH domain only 1 |
| chr17_+_67957848 | 0.08 |
ENST00000455460.1
|
AC004562.1
|
AC004562.1 |
| chr2_+_173955327 | 0.08 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr3_+_150264458 | 0.08 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr11_-_122930121 | 0.08 |
ENST00000524552.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr17_-_40169161 | 0.08 |
ENST00000589586.2
ENST00000426588.3 ENST00000589576.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr5_+_43602750 | 0.08 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr1_+_209878182 | 0.08 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr14_-_75330537 | 0.08 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr11_-_123185475 | 0.08 |
ENST00000527774.1
ENST00000527533.1 |
RP11-109E10.1
|
RP11-109E10.1 |
| chr1_+_61547405 | 0.08 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr19_-_10446449 | 0.08 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr12_+_56114151 | 0.08 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr11_-_59383617 | 0.08 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr2_-_211036051 | 0.08 |
ENST00000418791.1
ENST00000452086.1 ENST00000281772.9 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like |
| chr10_-_75193308 | 0.08 |
ENST00000299432.2
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr16_-_67185117 | 0.08 |
ENST00000449549.3
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr9_-_32635667 | 0.08 |
ENST00000242310.4
|
TAF1L
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 210kDa-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFIA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.2 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.4 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.3 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.3 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.4 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.0 | 0.2 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:2000005 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.2 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.3 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 0.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.3 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 1.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.0 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |