Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
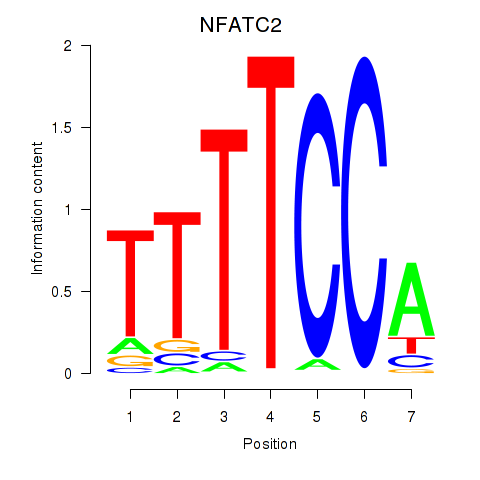
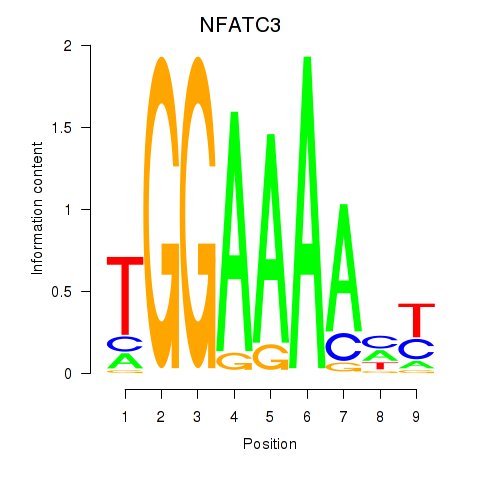
Results for NFATC2_NFATC3
Z-value: 1.21
Transcription factors associated with NFATC2_NFATC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC2
|
ENSG00000101096.15 | nuclear factor of activated T cells 2 |
|
NFATC3
|
ENSG00000072736.14 | nuclear factor of activated T cells 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC3 | hg19_v2_chr16_+_68119440_68119561 | 0.67 | 1.5e-01 | Click! |
| NFATC2 | hg19_v2_chr20_-_50179368_50179392 | 0.16 | 7.7e-01 | Click! |
Activity profile of NFATC2_NFATC3 motif
Sorted Z-values of NFATC2_NFATC3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_85969774 | 1.76 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr2_+_20650796 | 1.42 |
ENST00000448241.1
|
AC023137.2
|
AC023137.2 |
| chrX_-_15683147 | 1.14 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr6_+_148593425 | 0.87 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr6_-_13290684 | 0.80 |
ENST00000606393.1
|
RP1-257A7.5
|
RP1-257A7.5 |
| chr14_+_32476072 | 0.76 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chrX_+_152683780 | 0.65 |
ENST00000338647.5
|
ZFP92
|
ZFP92 zinc finger protein |
| chr19_+_36346734 | 0.63 |
ENST00000586102.3
|
KIRREL2
|
kin of IRRE like 2 (Drosophila) |
| chr5_+_35852797 | 0.62 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr17_+_6900201 | 0.55 |
ENST00000480801.1
|
ALOX12
|
arachidonate 12-lipoxygenase |
| chr5_-_111093759 | 0.54 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr14_-_51863853 | 0.53 |
ENST00000556762.1
|
RP11-255G12.3
|
RP11-255G12.3 |
| chr10_-_16563870 | 0.53 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr1_+_209941942 | 0.52 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr6_+_144665237 | 0.50 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr6_+_26183958 | 0.47 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr5_-_78808617 | 0.47 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr3_-_111852061 | 0.44 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr14_-_77889860 | 0.42 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr17_-_15469590 | 0.42 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr6_+_31105426 | 0.41 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr2_+_120517717 | 0.41 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr9_+_35806082 | 0.40 |
ENST00000447210.1
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr17_+_44701402 | 0.40 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr7_-_139756791 | 0.39 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr12_-_109747023 | 0.38 |
ENST00000355216.1
ENST00000299162.5 |
FOXN4
|
forkhead box N4 |
| chr7_+_28452130 | 0.38 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr15_+_93443419 | 0.37 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr5_+_140480083 | 0.37 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr15_+_84841242 | 0.37 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr5_-_108063949 | 0.36 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr22_-_28490123 | 0.35 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr1_-_151319318 | 0.35 |
ENST00000436271.1
ENST00000450506.1 ENST00000422595.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr20_+_62369623 | 0.35 |
ENST00000467211.1
|
RP4-583P15.14
|
RP4-583P15.14 |
| chr5_+_131993856 | 0.35 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr11_+_101983176 | 0.34 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr9_-_13165457 | 0.34 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr6_+_106535455 | 0.34 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr3_+_69812877 | 0.33 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr3_-_161089289 | 0.33 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr11_-_75062730 | 0.33 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr1_+_36038971 | 0.33 |
ENST00000373235.3
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr9_-_73483926 | 0.33 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr22_+_45680822 | 0.32 |
ENST00000216211.4
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr16_+_86612112 | 0.32 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr13_-_46716969 | 0.32 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_+_6554021 | 0.32 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr18_-_33047039 | 0.32 |
ENST00000591141.1
ENST00000586741.1 |
RP11-322E11.5
|
RP11-322E11.5 |
| chr9_+_129097520 | 0.31 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr12_-_52585765 | 0.31 |
ENST00000313234.5
ENST00000394815.2 |
KRT80
|
keratin 80 |
| chr6_+_31916733 | 0.30 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chrX_+_133941218 | 0.30 |
ENST00000370784.4
ENST00000370785.3 |
FAM122C
|
family with sequence similarity 122C |
| chr11_-_75062829 | 0.30 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr1_+_81106951 | 0.29 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr6_+_138188351 | 0.29 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr7_-_41742697 | 0.29 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr17_-_28661065 | 0.29 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr14_-_76447336 | 0.29 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor, beta 3 |
| chr6_-_15586238 | 0.28 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr11_-_57089774 | 0.28 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr2_+_198570081 | 0.27 |
ENST00000282276.6
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr15_-_44069513 | 0.27 |
ENST00000433927.1
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr18_+_43405471 | 0.27 |
ENST00000389474.3
|
SIGLEC15
|
sialic acid binding Ig-like lectin 15 |
| chr2_+_177028805 | 0.27 |
ENST00000249440.3
|
HOXD3
|
homeobox D3 |
| chr6_+_41021027 | 0.27 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr2_+_234580499 | 0.26 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_-_40657397 | 0.26 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr6_-_136788001 | 0.26 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr12_+_79357815 | 0.26 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr3_-_195310802 | 0.26 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr14_-_54418598 | 0.26 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr12_-_88423164 | 0.26 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr5_+_61708488 | 0.25 |
ENST00000505902.1
|
IPO11
|
importin 11 |
| chr14_+_77582905 | 0.25 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr5_-_124081008 | 0.25 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr9_+_19049372 | 0.25 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr6_-_30685214 | 0.25 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr9_-_136283156 | 0.25 |
ENST00000371942.3
|
REXO4
|
REX4, RNA exonuclease 4 homolog (S. cerevisiae) |
| chr14_-_23479331 | 0.24 |
ENST00000397377.1
ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr4_-_184243561 | 0.24 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr17_+_79953310 | 0.24 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr16_-_10868853 | 0.24 |
ENST00000572428.1
|
TVP23A
|
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
| chrX_-_154255143 | 0.23 |
ENST00000453950.1
ENST00000423959.1 |
F8
|
coagulation factor VIII, procoagulant component |
| chr2_+_232316906 | 0.23 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr5_+_74011328 | 0.23 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr18_-_24237339 | 0.23 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr11_+_18417948 | 0.23 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr2_-_190044480 | 0.23 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr4_-_140477353 | 0.22 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_-_111424462 | 0.22 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr14_+_77642191 | 0.22 |
ENST00000555338.1
ENST00000556514.1 |
TMEM63C
|
transmembrane protein 63C |
| chr1_+_247579451 | 0.22 |
ENST00000391828.3
ENST00000366497.2 |
NLRP3
|
NLR family, pyrin domain containing 3 |
| chr6_-_10415218 | 0.22 |
ENST00000466073.1
ENST00000498450.1 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr5_-_64777685 | 0.22 |
ENST00000536360.1
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr15_+_96869165 | 0.22 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr11_-_119293903 | 0.21 |
ENST00000580275.1
|
THY1
|
Thy-1 cell surface antigen |
| chr8_-_13134045 | 0.21 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr2_+_234580525 | 0.21 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_239882842 | 0.21 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr17_+_59477233 | 0.21 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr17_-_39041479 | 0.21 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr5_-_131879205 | 0.21 |
ENST00000231454.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr11_+_34663913 | 0.21 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr14_+_38065052 | 0.21 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr17_-_15522826 | 0.21 |
ENST00000395906.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr12_-_67197760 | 0.21 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr11_-_123065989 | 0.21 |
ENST00000448775.2
|
CLMP
|
CXADR-like membrane protein |
| chr11_+_123986069 | 0.20 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr15_+_77713299 | 0.20 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr14_+_78227105 | 0.20 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr2_+_191334212 | 0.20 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr1_-_27226928 | 0.20 |
ENST00000361720.5
|
GPATCH3
|
G patch domain containing 3 |
| chr1_+_86046433 | 0.20 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr5_-_146461027 | 0.20 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr4_-_71532339 | 0.20 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr5_+_137722255 | 0.20 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr1_+_206623784 | 0.20 |
ENST00000426388.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr2_-_37415690 | 0.20 |
ENST00000260637.3
ENST00000535679.1 ENST00000379149.2 |
SULT6B1
|
sulfotransferase family, cytosolic, 6B, member 1 |
| chr8_+_94712895 | 0.20 |
ENST00000523453.1
ENST00000520955.1 |
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr11_-_119187826 | 0.19 |
ENST00000264036.4
|
MCAM
|
melanoma cell adhesion molecule |
| chr15_+_32933866 | 0.19 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr11_+_9482512 | 0.19 |
ENST00000396602.2
ENST00000530463.1 ENST00000533542.1 ENST00000532577.1 ENST00000396597.3 |
ZNF143
|
zinc finger protein 143 |
| chr10_-_101825151 | 0.19 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr6_-_26235206 | 0.19 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr20_+_58179582 | 0.19 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr5_+_140186647 | 0.19 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr14_-_88200641 | 0.19 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr5_-_34043310 | 0.19 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr18_+_61442629 | 0.19 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr16_-_1993124 | 0.19 |
ENST00000473663.1
ENST00000399753.2 ENST00000564908.1 |
MSRB1
|
methionine sulfoxide reductase B1 |
| chr20_-_36156264 | 0.19 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr9_+_133589333 | 0.19 |
ENST00000372348.2
ENST00000393293.4 |
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chrX_-_41782592 | 0.18 |
ENST00000378158.1
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr8_+_92114873 | 0.18 |
ENST00000343709.3
ENST00000448384.2 |
LRRC69
|
leucine rich repeat containing 69 |
| chr17_-_61045902 | 0.18 |
ENST00000581596.1
|
RP11-180P8.3
|
RP11-180P8.3 |
| chr17_-_47045949 | 0.18 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr12_+_96588368 | 0.18 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr18_+_3451584 | 0.18 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr12_+_26348582 | 0.18 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr5_-_36301984 | 0.18 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr17_-_38256973 | 0.18 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr12_-_50616382 | 0.17 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr13_-_81801115 | 0.17 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr16_-_3306587 | 0.17 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr11_-_8892464 | 0.17 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr17_+_79495397 | 0.17 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr19_+_41281060 | 0.17 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr9_-_13175823 | 0.17 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr2_-_228244013 | 0.17 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr5_+_127039075 | 0.17 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr12_-_50616122 | 0.17 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr3_-_156878482 | 0.17 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr13_-_33924755 | 0.17 |
ENST00000439831.1
ENST00000567873.1 |
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr12_-_21910775 | 0.17 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr11_+_13299186 | 0.16 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr7_+_22766766 | 0.16 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chrX_+_9431324 | 0.16 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr1_+_67632083 | 0.16 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr14_+_21569245 | 0.16 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr5_-_124082279 | 0.16 |
ENST00000513986.1
|
ZNF608
|
zinc finger protein 608 |
| chr5_+_167718604 | 0.16 |
ENST00000265293.4
|
WWC1
|
WW and C2 domain containing 1 |
| chr2_+_136343904 | 0.16 |
ENST00000436436.1
|
R3HDM1
|
R3H domain containing 1 |
| chr11_-_72492903 | 0.16 |
ENST00000537947.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr6_+_106534192 | 0.16 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr8_+_128748466 | 0.16 |
ENST00000524013.1
ENST00000520751.1 |
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr2_-_191885686 | 0.16 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr18_-_25739260 | 0.16 |
ENST00000413878.1
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr2_-_207082748 | 0.16 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr14_-_103589246 | 0.16 |
ENST00000558224.1
ENST00000560742.1 |
LINC00677
|
long intergenic non-protein coding RNA 677 |
| chr3_-_100565249 | 0.15 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_-_68698197 | 0.15 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr17_+_20059358 | 0.15 |
ENST00000536879.1
ENST00000395522.2 ENST00000395525.3 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr3_+_111578131 | 0.15 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_+_26205496 | 0.15 |
ENST00000537946.1
ENST00000541218.1 ENST00000282884.9 ENST00000545413.1 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr6_+_144606817 | 0.15 |
ENST00000433557.1
|
UTRN
|
utrophin |
| chr12_-_122240792 | 0.15 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr17_-_26127525 | 0.15 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr13_-_33780133 | 0.15 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr16_+_67280799 | 0.15 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr22_-_29107919 | 0.15 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr9_-_16705069 | 0.15 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr3_-_124839648 | 0.15 |
ENST00000430155.2
|
SLC12A8
|
solute carrier family 12, member 8 |
| chr18_-_24129367 | 0.15 |
ENST00000408011.3
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr3_+_140981456 | 0.15 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr1_-_156675535 | 0.15 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_+_18647326 | 0.15 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr17_-_36981556 | 0.15 |
ENST00000536127.1
ENST00000225428.5 |
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chrX_+_18725758 | 0.14 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr11_-_62457371 | 0.14 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr5_+_38148582 | 0.14 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr5_+_59783941 | 0.14 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr2_+_162272605 | 0.14 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr14_-_25479811 | 0.14 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr11_-_46142948 | 0.14 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr11_+_57531292 | 0.14 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr17_-_46688334 | 0.14 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr15_-_44969086 | 0.14 |
ENST00000434130.1
ENST00000560780.1 |
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr12_+_113354341 | 0.14 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr22_-_36236265 | 0.14 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr11_+_111807863 | 0.14 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC2_NFATC3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 0.5 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.6 | GO:2001301 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.4 | GO:0070433 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.6 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.1 | 0.7 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 1.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.4 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.1 | 0.5 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 0.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.2 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.1 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.6 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.6 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0072101 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.3 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 0.2 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.4 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.2 | GO:0021622 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.2 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.2 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.3 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.7 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.3 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:1990828 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.3 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.2 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.3 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.2 | GO:2000809 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.0 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.3 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0038194 | thyroid-stimulating hormone signaling pathway(GO:0038194) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.0 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.0 | GO:0046086 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.4 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.0 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 1.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.6 | GO:0047977 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0034617 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |