Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
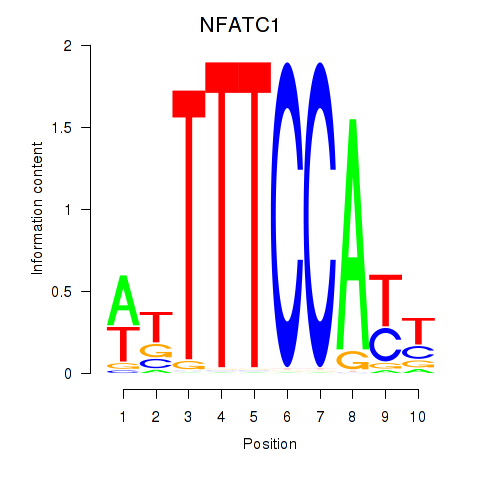
Results for NFATC1
Z-value: 1.00
Transcription factors associated with NFATC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC1
|
ENSG00000131196.13 | nuclear factor of activated T cells 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC1 | hg19_v2_chr18_+_77155856_77155939 | -0.76 | 8.1e-02 | Click! |
Activity profile of NFATC1 motif
Sorted Z-values of NFATC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_94590910 | 1.07 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr1_+_948803 | 1.05 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr12_-_92536433 | 0.96 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr15_-_35280426 | 0.72 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr15_+_38964048 | 0.70 |
ENST00000560203.1
ENST00000557946.1 |
RP11-275I4.2
|
RP11-275I4.2 |
| chrX_-_84634737 | 0.65 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr1_+_79115503 | 0.62 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr5_+_49962495 | 0.60 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr14_+_32476072 | 0.58 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr14_-_92413727 | 0.58 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr2_+_46926326 | 0.56 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr14_-_92413353 | 0.53 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr11_+_34642656 | 0.52 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr10_+_22610124 | 0.47 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr2_+_33701707 | 0.45 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr21_-_16374688 | 0.45 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr2_-_42160486 | 0.44 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr2_+_46926048 | 0.44 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr3_+_88188254 | 0.43 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr1_+_87595433 | 0.42 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr5_-_41510725 | 0.41 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr12_+_20968608 | 0.41 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr4_+_86748898 | 0.41 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_-_178976996 | 0.41 |
ENST00000485523.1
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr1_+_150254936 | 0.41 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr9_-_19065082 | 0.40 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr6_+_135818979 | 0.39 |
ENST00000421378.2
ENST00000579057.1 ENST00000436554.1 ENST00000438618.2 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr16_+_53412368 | 0.38 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr1_-_186649543 | 0.37 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chrX_-_84634708 | 0.36 |
ENST00000373145.3
|
POF1B
|
premature ovarian failure, 1B |
| chr11_-_110167331 | 0.36 |
ENST00000534683.1
|
RDX
|
radixin |
| chr2_+_33701684 | 0.35 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr13_-_49975632 | 0.35 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr2_+_201390843 | 0.34 |
ENST00000357799.4
ENST00000409203.3 |
SGOL2
|
shugoshin-like 2 (S. pombe) |
| chr13_+_37581115 | 0.34 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr5_+_118690466 | 0.33 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr9_+_22646189 | 0.33 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr14_-_35591433 | 0.33 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr12_-_13529594 | 0.32 |
ENST00000539026.1
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr11_-_110167352 | 0.32 |
ENST00000533991.1
ENST00000528498.1 ENST00000405097.1 ENST00000528900.1 ENST00000530301.1 ENST00000343115.4 |
RDX
|
radixin |
| chr3_+_152017360 | 0.31 |
ENST00000485910.1
ENST00000463374.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr7_-_87505658 | 0.31 |
ENST00000341119.5
|
SLC25A40
|
solute carrier family 25, member 40 |
| chr5_-_78808617 | 0.31 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr14_+_32547434 | 0.31 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr15_-_30686052 | 0.30 |
ENST00000562729.1
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr5_-_39203093 | 0.30 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr2_-_228244013 | 0.30 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr5_+_118691008 | 0.30 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr13_-_108867101 | 0.30 |
ENST00000356922.4
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr13_+_96085847 | 0.30 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr10_+_43932553 | 0.29 |
ENST00000456416.1
ENST00000437590.2 ENST00000451167.1 |
ZNF487
|
zinc finger protein 487 |
| chr11_+_130542851 | 0.29 |
ENST00000317019.2
|
C11orf44
|
chromosome 11 open reading frame 44 |
| chr1_+_92632542 | 0.29 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr11_+_11863500 | 0.29 |
ENST00000527733.1
ENST00000539466.1 |
USP47
|
ubiquitin specific peptidase 47 |
| chr17_+_79495397 | 0.29 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr1_+_167298281 | 0.29 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr11_+_36616044 | 0.29 |
ENST00000334307.5
ENST00000531554.1 ENST00000347206.4 ENST00000534635.1 ENST00000446510.2 ENST00000530697.1 ENST00000527108.1 |
C11orf74
|
chromosome 11 open reading frame 74 |
| chr7_-_127032114 | 0.29 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr3_+_69134124 | 0.28 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr1_+_235491714 | 0.28 |
ENST00000471812.1
ENST00000358966.2 ENST00000282841.5 ENST00000391855.2 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr11_+_11863579 | 0.28 |
ENST00000399455.2
|
USP47
|
ubiquitin specific peptidase 47 |
| chr21_-_35899113 | 0.26 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr20_+_36149602 | 0.26 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr14_+_23025534 | 0.26 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr5_-_41510656 | 0.26 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr12_+_26111823 | 0.26 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr2_-_179914760 | 0.26 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr19_+_35939154 | 0.26 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr1_-_94703118 | 0.25 |
ENST00000260526.6
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr13_-_67804445 | 0.25 |
ENST00000456367.1
ENST00000377861.3 ENST00000544246.1 |
PCDH9
|
protocadherin 9 |
| chr8_+_86121448 | 0.25 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr7_-_151217166 | 0.25 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr14_+_90864504 | 0.25 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr14_-_35591156 | 0.24 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr4_+_86749045 | 0.24 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr7_-_103848405 | 0.24 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr1_-_153517473 | 0.24 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr12_+_52431016 | 0.24 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr5_+_32788945 | 0.24 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr15_-_59981479 | 0.24 |
ENST00000607373.1
|
BNIP2
|
BCL2/adenovirus E1B 19kDa interacting protein 2 |
| chr5_+_41904431 | 0.24 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr7_-_83824449 | 0.23 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr18_+_61575200 | 0.23 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr12_+_69201923 | 0.23 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr8_+_38831683 | 0.23 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr10_-_65028817 | 0.22 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr12_-_90103077 | 0.22 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr7_-_105319536 | 0.22 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr15_+_23255242 | 0.22 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr14_+_58894706 | 0.22 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chrX_+_102611373 | 0.22 |
ENST00000372661.3
ENST00000372656.3 |
WBP5
|
WW domain binding protein 5 |
| chr18_+_77155942 | 0.21 |
ENST00000397790.2
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr2_-_172087824 | 0.21 |
ENST00000521943.1
|
TLK1
|
tousled-like kinase 1 |
| chr19_-_44388116 | 0.21 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr4_-_38806404 | 0.21 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr3_+_107364769 | 0.21 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_-_50889155 | 0.21 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr19_+_10197463 | 0.21 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr8_+_92082424 | 0.20 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chr9_-_20621834 | 0.20 |
ENST00000429426.2
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr5_+_33441053 | 0.20 |
ENST00000541634.1
ENST00000455217.2 ENST00000414361.2 |
TARS
|
threonyl-tRNA synthetase |
| chrX_-_119763835 | 0.20 |
ENST00000371313.2
ENST00000304661.5 |
C1GALT1C1
|
C1GALT1-specific chaperone 1 |
| chr4_-_71532668 | 0.20 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr10_-_65028938 | 0.20 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr12_-_16760021 | 0.20 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_-_95449155 | 0.19 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr7_-_6010263 | 0.19 |
ENST00000455618.2
ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr3_+_184530173 | 0.19 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr1_-_77685084 | 0.19 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr21_-_35340759 | 0.19 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr3_-_158390282 | 0.19 |
ENST00000264265.3
|
LXN
|
latexin |
| chr14_+_51026926 | 0.19 |
ENST00000557735.1
|
ATL1
|
atlastin GTPase 1 |
| chr15_-_35261996 | 0.19 |
ENST00000156471.5
|
AQR
|
aquarius intron-binding spliceosomal factor |
| chr11_-_102401469 | 0.18 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr1_-_111743285 | 0.18 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr3_+_133293278 | 0.18 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr5_+_7869217 | 0.18 |
ENST00000264668.2
ENST00000514220.1 ENST00000341013.6 ENST00000440940.2 ENST00000502550.1 ENST00000506877.1 |
MTRR
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr3_+_107364683 | 0.18 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr2_-_68479614 | 0.18 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr8_-_17555164 | 0.18 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr4_+_39046615 | 0.18 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr7_-_100493744 | 0.18 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr17_+_27047570 | 0.18 |
ENST00000472628.1
ENST00000578181.1 |
RPL23A
|
ribosomal protein L23a |
| chr1_+_186649754 | 0.18 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr3_+_148447887 | 0.17 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr1_+_218519577 | 0.17 |
ENST00000366930.4
ENST00000366929.4 |
TGFB2
|
transforming growth factor, beta 2 |
| chr14_+_35591509 | 0.17 |
ENST00000604073.1
|
KIAA0391
|
KIAA0391 |
| chr4_-_164534657 | 0.17 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr4_-_153274078 | 0.17 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr5_+_33440802 | 0.17 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr16_+_68573116 | 0.16 |
ENST00000570495.1
ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr5_+_75904950 | 0.16 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr5_+_135496675 | 0.16 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr14_+_35591020 | 0.16 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr11_-_13461790 | 0.16 |
ENST00000530907.1
|
BTBD10
|
BTB (POZ) domain containing 10 |
| chr2_+_86668464 | 0.16 |
ENST00000409064.1
|
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr3_+_130648842 | 0.16 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr12_+_104359641 | 0.16 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr1_-_85462762 | 0.16 |
ENST00000284027.5
|
MCOLN2
|
mucolipin 2 |
| chr15_+_63354769 | 0.16 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr2_-_40679148 | 0.16 |
ENST00000417271.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr11_-_122929699 | 0.15 |
ENST00000526686.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr8_-_102987548 | 0.15 |
ENST00000520690.1
|
NCALD
|
neurocalcin delta |
| chr3_+_141596371 | 0.15 |
ENST00000495216.1
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr4_-_168155300 | 0.15 |
ENST00000541637.1
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_+_22351977 | 0.15 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr19_-_30199516 | 0.15 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr6_-_56716686 | 0.15 |
ENST00000520645.1
|
DST
|
dystonin |
| chr17_+_49337881 | 0.15 |
ENST00000225298.7
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast) |
| chr8_+_57124245 | 0.15 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr3_+_178866199 | 0.15 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr1_-_85462623 | 0.15 |
ENST00000370608.3
|
MCOLN2
|
mucolipin 2 |
| chr17_+_47287749 | 0.15 |
ENST00000419580.2
|
ABI3
|
ABI family, member 3 |
| chr1_+_82266053 | 0.14 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr3_+_157261116 | 0.14 |
ENST00000468043.1
ENST00000459838.1 ENST00000461040.1 ENST00000449199.2 ENST00000426338.2 |
C3orf55
|
chromosome 3 open reading frame 55 |
| chr6_+_22221010 | 0.14 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr16_-_85969774 | 0.14 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr1_+_61548225 | 0.14 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr3_+_69134080 | 0.14 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr4_-_104020968 | 0.14 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr12_-_56352368 | 0.14 |
ENST00000549404.1
|
PMEL
|
premelanosome protein |
| chr12_+_104359614 | 0.14 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr7_+_107220422 | 0.14 |
ENST00000005259.4
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr5_-_7869108 | 0.14 |
ENST00000264669.5
ENST00000507572.1 ENST00000504695.1 |
FASTKD3
|
FAST kinase domains 3 |
| chr3_-_27498235 | 0.14 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr14_+_81421921 | 0.14 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chr11_+_18417813 | 0.14 |
ENST00000540430.1
ENST00000379412.5 |
LDHA
|
lactate dehydrogenase A |
| chr12_-_7245018 | 0.14 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr2_-_230786032 | 0.14 |
ENST00000428959.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chrX_-_48827976 | 0.14 |
ENST00000218176.3
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr1_-_103574024 | 0.14 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr14_+_51026844 | 0.14 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr11_-_122930121 | 0.14 |
ENST00000524552.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr11_+_392587 | 0.14 |
ENST00000534401.1
|
PKP3
|
plakophilin 3 |
| chr1_-_91487013 | 0.13 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr12_+_131356582 | 0.13 |
ENST00000448750.3
ENST00000541630.1 ENST00000392369.2 ENST00000254675.3 ENST00000535090.1 ENST00000392367.3 |
RAN
|
RAN, member RAS oncogene family |
| chr17_-_46692457 | 0.13 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr16_-_66952742 | 0.13 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr6_-_130536774 | 0.13 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr1_+_203734296 | 0.13 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr8_+_81397846 | 0.13 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr14_+_51026743 | 0.13 |
ENST00000358385.6
ENST00000357032.3 ENST00000354525.4 |
ATL1
|
atlastin GTPase 1 |
| chr6_-_134495992 | 0.13 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr9_+_123906331 | 0.13 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr12_+_34175398 | 0.12 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr10_-_63995871 | 0.12 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr9_-_20622478 | 0.12 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr6_-_64029879 | 0.12 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr17_+_27046988 | 0.12 |
ENST00000496182.1
|
RPL23A
|
ribosomal protein L23a |
| chr4_-_155471528 | 0.12 |
ENST00000302078.5
ENST00000499023.2 |
PLRG1
|
pleiotropic regulator 1 |
| chr12_-_123756781 | 0.12 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr4_+_154387480 | 0.12 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr13_+_49822041 | 0.12 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr16_-_66952779 | 0.12 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr17_-_49021974 | 0.12 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr9_-_16253112 | 0.12 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr1_-_151798546 | 0.12 |
ENST00000356728.6
|
RORC
|
RAR-related orphan receptor C |
| chr3_-_136471204 | 0.11 |
ENST00000480733.1
ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1
|
stromal antigen 1 |
| chr21_+_35445811 | 0.11 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr10_-_27149904 | 0.11 |
ENST00000376166.1
ENST00000376138.3 ENST00000355394.4 ENST00000346832.5 ENST00000376134.3 ENST00000376137.4 ENST00000536334.1 ENST00000490841.2 |
ABI1
|
abl-interactor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.7 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 1.0 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.3 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.4 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.2 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.2 | GO:0070340 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.2 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.3 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 0.6 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 0.2 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) uterine wall breakdown(GO:0042704) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.4 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.8 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0038194 | thyroid-stimulating hormone signaling pathway(GO:0038194) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0046292 | spermatid nucleus elongation(GO:0007290) formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:1902871 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) sensory system development(GO:0048880) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.0 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.2 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.1 | GO:0098907 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.9 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.4 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.1 | 0.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |