Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
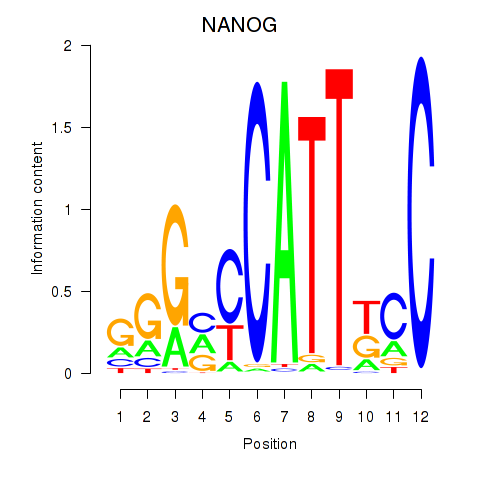
Results for NANOG
Z-value: 0.82
Transcription factors associated with NANOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NANOG
|
ENSG00000111704.6 | Nanog homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NANOG | hg19_v2_chr12_+_7941989_7942014 | 0.46 | 3.6e-01 | Click! |
Activity profile of NANOG motif
Sorted Z-values of NANOG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_53901266 | 0.81 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chrX_-_16888276 | 0.73 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr1_-_151254362 | 0.54 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr17_-_55911970 | 0.49 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr12_-_123187890 | 0.44 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr19_+_37825614 | 0.41 |
ENST00000591259.1
ENST00000590582.1 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr12_-_100378392 | 0.41 |
ENST00000549866.1
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_+_59486129 | 0.41 |
ENST00000438195.1
ENST00000424308.1 |
RP4-794H19.4
|
RP4-794H19.4 |
| chr12_+_57828521 | 0.41 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr6_+_26087646 | 0.37 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr3_-_52567792 | 0.34 |
ENST00000307092.4
ENST00000422318.2 ENST00000459839.1 |
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr9_+_131902346 | 0.31 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr6_+_26087509 | 0.30 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr1_-_145589424 | 0.27 |
ENST00000334513.5
|
NUDT17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr6_+_292253 | 0.27 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr2_-_97536490 | 0.26 |
ENST00000449330.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr4_+_154073469 | 0.25 |
ENST00000441616.1
|
TRIM2
|
tripartite motif containing 2 |
| chr6_-_31633402 | 0.24 |
ENST00000375893.2
|
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr4_+_3344141 | 0.24 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr19_-_39926268 | 0.24 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr12_+_25055243 | 0.23 |
ENST00000599478.1
|
AC026310.1
|
Protein 101060047 |
| chr17_+_7387677 | 0.22 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr14_+_101295948 | 0.21 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr5_+_72509751 | 0.21 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr19_-_13213662 | 0.21 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr8_-_42751728 | 0.21 |
ENST00000319104.3
ENST00000531440.1 |
RNF170
|
ring finger protein 170 |
| chr3_-_45837959 | 0.21 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr6_+_31540056 | 0.20 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr2_+_87769459 | 0.19 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr16_+_31494323 | 0.19 |
ENST00000569576.1
ENST00000330498.3 |
SLC5A2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr13_+_108921977 | 0.19 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr14_-_98444438 | 0.19 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr15_+_90319557 | 0.18 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr6_+_292459 | 0.18 |
ENST00000419235.2
ENST00000605035.1 ENST00000605863.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr11_+_1295809 | 0.17 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr12_-_42878101 | 0.17 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr10_+_123923205 | 0.17 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr5_-_152069089 | 0.17 |
ENST00000506723.2
|
AC091969.1
|
AC091969.1 |
| chr1_+_44870866 | 0.17 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr17_-_43128943 | 0.16 |
ENST00000588499.1
ENST00000593094.1 |
DCAKD
|
dephospho-CoA kinase domain containing |
| chr19_+_42381173 | 0.16 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr10_-_1034237 | 0.16 |
ENST00000381466.1
|
AL359878.1
|
Uncharacterized protein |
| chr1_-_150241341 | 0.16 |
ENST00000369109.3
ENST00000414276.2 ENST00000236017.5 |
APH1A
|
APH1A gamma secretase subunit |
| chr3_-_71803917 | 0.16 |
ENST00000421769.2
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr16_+_14280742 | 0.16 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr17_-_61777459 | 0.16 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr10_-_70092635 | 0.16 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr9_+_131902283 | 0.15 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr19_+_39926791 | 0.15 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr4_-_140544386 | 0.15 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr6_+_32407619 | 0.15 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr2_+_171673072 | 0.15 |
ENST00000358196.3
ENST00000375272.1 |
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr6_-_43021437 | 0.14 |
ENST00000265348.3
|
CUL7
|
cullin 7 |
| chr16_+_3507985 | 0.14 |
ENST00000421765.3
ENST00000360862.5 ENST00000414063.2 ENST00000610180.1 ENST00000608993.1 |
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr5_-_33892204 | 0.14 |
ENST00000504830.1
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr4_+_114038137 | 0.13 |
ENST00000508613.1
|
ANK2
|
ankyrin 2, neuronal |
| chr7_+_150382781 | 0.13 |
ENST00000223293.5
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr19_+_41816053 | 0.13 |
ENST00000269967.3
|
CCDC97
|
coiled-coil domain containing 97 |
| chr19_-_39926555 | 0.13 |
ENST00000599539.1
ENST00000339471.4 ENST00000601655.1 ENST00000251453.3 |
RPS16
|
ribosomal protein S16 |
| chr2_-_214016314 | 0.13 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr10_-_70092671 | 0.13 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr10_+_123922941 | 0.13 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_+_22022800 | 0.12 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr12_-_69080590 | 0.12 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr19_+_4969116 | 0.12 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr1_+_27719148 | 0.12 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr19_-_39342962 | 0.12 |
ENST00000600873.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr7_+_144015218 | 0.12 |
ENST00000408951.1
|
OR2A1
|
olfactory receptor, family 2, subfamily A, member 1 |
| chr12_-_123215306 | 0.12 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr22_-_42084863 | 0.11 |
ENST00000401959.1
ENST00000355257.3 |
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr3_+_128968437 | 0.11 |
ENST00000314797.6
|
COPG1
|
coatomer protein complex, subunit gamma 1 |
| chr10_-_49812997 | 0.11 |
ENST00000417912.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr1_+_59486059 | 0.11 |
ENST00000447329.1
|
RP4-794H19.4
|
RP4-794H19.4 |
| chr17_-_72542278 | 0.10 |
ENST00000330793.1
|
CD300C
|
CD300c molecule |
| chr6_-_43484718 | 0.10 |
ENST00000372422.2
|
YIPF3
|
Yip1 domain family, member 3 |
| chr6_+_148663729 | 0.10 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr11_+_126173647 | 0.10 |
ENST00000263579.4
|
DCPS
|
decapping enzyme, scavenger |
| chr10_-_49813090 | 0.10 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chrX_-_107019181 | 0.10 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr5_+_156693091 | 0.09 |
ENST00000318218.6
ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_-_23670546 | 0.09 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr6_-_43021612 | 0.09 |
ENST00000535468.1
|
CUL7
|
cullin 7 |
| chr1_+_203097407 | 0.09 |
ENST00000367235.1
|
ADORA1
|
adenosine A1 receptor |
| chr19_-_42806444 | 0.09 |
ENST00000594989.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr11_-_72504637 | 0.09 |
ENST00000536377.1
ENST00000359373.5 |
STARD10
ARAP1
|
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr11_-_72504681 | 0.09 |
ENST00000538536.1
ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr3_-_53925863 | 0.09 |
ENST00000541726.1
ENST00000495461.1 |
SELK
|
Selenoprotein K |
| chr3_+_38388251 | 0.09 |
ENST00000427323.1
ENST00000207870.3 ENST00000542835.1 |
XYLB
|
xylulokinase homolog (H. influenzae) |
| chr6_+_43484760 | 0.08 |
ENST00000372389.3
ENST00000372344.2 ENST00000304004.3 ENST00000423780.1 |
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa |
| chr16_+_30759700 | 0.08 |
ENST00000328273.7
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr16_+_28986134 | 0.08 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr10_+_123923105 | 0.08 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chrX_+_47082408 | 0.08 |
ENST00000518022.1
ENST00000276052.6 |
CDK16
|
cyclin-dependent kinase 16 |
| chr16_+_30759563 | 0.07 |
ENST00000563588.1
ENST00000565924.1 ENST00000424889.3 |
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr4_+_166248775 | 0.07 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr7_+_131012605 | 0.07 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr15_+_77713222 | 0.07 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr2_-_240322685 | 0.07 |
ENST00000544989.1
|
HDAC4
|
histone deacetylase 4 |
| chr10_-_79686284 | 0.07 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr6_+_52226897 | 0.07 |
ENST00000442253.2
|
PAQR8
|
progestin and adipoQ receptor family member VIII |
| chr12_-_101604185 | 0.07 |
ENST00000536262.2
|
SLC5A8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chrX_+_14547632 | 0.07 |
ENST00000218075.4
|
GLRA2
|
glycine receptor, alpha 2 |
| chr11_+_63870660 | 0.07 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr2_-_240322643 | 0.07 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr8_-_27468842 | 0.07 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr8_+_9413410 | 0.07 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr12_-_46663734 | 0.07 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr19_-_50370509 | 0.07 |
ENST00000596014.1
|
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr19_+_8455077 | 0.07 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr19_-_42806723 | 0.06 |
ENST00000262890.3
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr17_+_75372165 | 0.06 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr7_-_2883928 | 0.06 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr8_+_22022653 | 0.06 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr6_-_43484621 | 0.06 |
ENST00000506469.1
ENST00000503972.1 |
YIPF3
|
Yip1 domain family, member 3 |
| chr5_+_156693159 | 0.06 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr8_-_27468945 | 0.06 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr16_-_2264779 | 0.06 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr17_-_7387524 | 0.06 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr14_+_23791159 | 0.05 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr16_-_75301886 | 0.05 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr3_+_15468862 | 0.05 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chrX_+_103173457 | 0.05 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B |
| chr9_+_124030338 | 0.05 |
ENST00000449773.1
ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN
|
gelsolin |
| chr2_+_219081817 | 0.05 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr1_+_151253991 | 0.05 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr11_+_3968573 | 0.05 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr19_-_8070474 | 0.05 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chrX_+_47092314 | 0.05 |
ENST00000218348.3
|
USP11
|
ubiquitin specific peptidase 11 |
| chr20_-_61557821 | 0.05 |
ENST00000354665.4
ENST00000370368.1 ENST00000395343.1 ENST00000395340.1 |
DIDO1
|
death inducer-obliterator 1 |
| chr14_-_23058063 | 0.04 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.4 |
DAD1
|
defender against cell death 1 |
| chr21_-_35340759 | 0.04 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr1_+_161551101 | 0.04 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr8_+_42752053 | 0.04 |
ENST00000307602.4
|
HOOK3
|
hook microtubule-tethering protein 3 |
| chr14_+_21156915 | 0.04 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr1_+_145575227 | 0.04 |
ENST00000393046.2
ENST00000369299.3 ENST00000463514.2 |
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr7_-_28220354 | 0.04 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr6_+_90272488 | 0.04 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr1_+_161632937 | 0.04 |
ENST00000236937.9
ENST00000367961.4 ENST00000358671.5 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr16_+_30759844 | 0.04 |
ENST00000565897.1
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr1_+_210406121 | 0.04 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr9_+_71819927 | 0.04 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr5_+_139493665 | 0.04 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr1_-_92952433 | 0.04 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr6_+_88032299 | 0.04 |
ENST00000608353.1
ENST00000392863.1 ENST00000229570.5 ENST00000608525.1 ENST00000608868.1 |
SMIM8
|
small integral membrane protein 8 |
| chr15_+_41549105 | 0.03 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr22_+_37447771 | 0.03 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr17_-_56065540 | 0.03 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr22_+_41487711 | 0.03 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr16_+_3508063 | 0.03 |
ENST00000576787.1
ENST00000572942.1 ENST00000576916.1 ENST00000575076.1 ENST00000572131.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr2_-_97405775 | 0.03 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr18_+_13218769 | 0.03 |
ENST00000399848.3
ENST00000361205.4 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr18_-_23671139 | 0.03 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr19_-_50370799 | 0.03 |
ENST00000600910.1
ENST00000322344.3 ENST00000600573.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr12_-_25055177 | 0.03 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr19_+_8455200 | 0.03 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr15_+_44580955 | 0.03 |
ENST00000345795.2
ENST00000360824.3 |
CASC4
|
cancer susceptibility candidate 4 |
| chr1_+_145575980 | 0.03 |
ENST00000393045.2
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr4_+_154074217 | 0.03 |
ENST00000437508.2
|
TRIM2
|
tripartite motif containing 2 |
| chr6_+_292051 | 0.03 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr2_-_112237835 | 0.03 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr7_-_2883650 | 0.03 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr8_-_27469196 | 0.03 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr7_-_76829125 | 0.03 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chr1_+_145576007 | 0.03 |
ENST00000369298.1
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr3_-_155572164 | 0.03 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr10_-_73497581 | 0.03 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr3_-_149093499 | 0.02 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr10_-_48390973 | 0.02 |
ENST00000224600.4
|
RBP3
|
retinol binding protein 3, interstitial |
| chr22_-_32026810 | 0.02 |
ENST00000266095.5
ENST00000397500.1 |
PISD
|
phosphatidylserine decarboxylase |
| chr6_+_34204642 | 0.02 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr4_+_79567314 | 0.02 |
ENST00000503539.1
ENST00000504675.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr5_-_115872124 | 0.02 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr13_-_103389159 | 0.02 |
ENST00000322527.2
|
CCDC168
|
coiled-coil domain containing 168 |
| chr9_+_71820057 | 0.02 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr11_+_2421718 | 0.02 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr1_+_36348790 | 0.01 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr10_+_1034646 | 0.01 |
ENST00000360059.5
ENST00000545048.1 |
GTPBP4
|
GTP binding protein 4 |
| chr6_+_30524663 | 0.01 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr1_-_155959853 | 0.01 |
ENST00000462460.2
ENST00000368316.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr15_-_50978965 | 0.01 |
ENST00000560955.1
ENST00000313478.7 |
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr18_+_3262954 | 0.01 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr6_-_33714752 | 0.01 |
ENST00000451316.1
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr14_+_101295638 | 0.01 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr6_+_155537771 | 0.01 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr17_-_56065484 | 0.01 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr12_-_123201337 | 0.00 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr10_-_118885954 | 0.00 |
ENST00000392901.4
|
KIAA1598
|
KIAA1598 |
| chr6_-_131277510 | 0.00 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_-_51297837 | 0.00 |
ENST00000245441.5
ENST00000389868.3 ENST00000382041.3 ENST00000324330.9 ENST00000453196.1 ENST00000453401.2 |
NIN
|
ninein (GSK3B interacting protein) |
| chr6_-_88032795 | 0.00 |
ENST00000296882.3
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr6_-_33714667 | 0.00 |
ENST00000293756.4
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr1_+_201924619 | 0.00 |
ENST00000367287.4
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NANOG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.2 | 0.8 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 0.7 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.2 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:2001112 | negative regulation of hepatocyte growth factor receptor signaling pathway(GO:1902203) regulation of cellular response to hepatocyte growth factor stimulus(GO:2001112) negative regulation of cellular response to hepatocyte growth factor stimulus(GO:2001113) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.2 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.2 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0043317 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.2 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.5 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.0 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |