Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
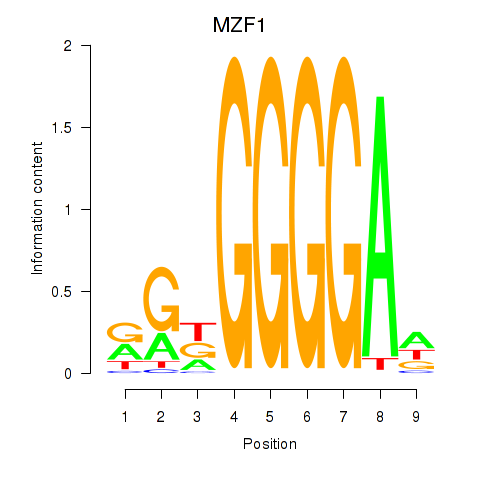
Results for MZF1
Z-value: 0.79
Transcription factors associated with MZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MZF1
|
ENSG00000099326.4 | myeloid zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MZF1 | hg19_v2_chr19_-_59084922_59084947 | 0.49 | 3.3e-01 | Click! |
Activity profile of MZF1 motif
Sorted Z-values of MZF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_45168875 | 0.59 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr9_-_140115775 | 0.40 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr14_+_29241910 | 0.38 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr3_+_119814070 | 0.35 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr9_+_116343192 | 0.34 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr22_+_40440804 | 0.29 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr17_+_7211656 | 0.27 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr17_+_7758374 | 0.25 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr17_+_55334364 | 0.24 |
ENST00000322684.3
ENST00000579590.1 |
MSI2
|
musashi RNA-binding protein 2 |
| chrX_-_45710920 | 0.24 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr2_+_206950095 | 0.23 |
ENST00000435627.1
|
AC007383.3
|
AC007383.3 |
| chr18_-_72920372 | 0.22 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr12_-_108991778 | 0.22 |
ENST00000549447.1
|
TMEM119
|
transmembrane protein 119 |
| chr11_-_46722117 | 0.22 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr3_-_178865747 | 0.21 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr17_+_7982800 | 0.21 |
ENST00000399413.3
|
AC129492.6
|
AC129492.6 |
| chr11_+_130184888 | 0.20 |
ENST00000602376.1
ENST00000532116.3 ENST00000602310.1 |
RP11-121M22.1
|
RP11-121M22.1 |
| chr17_-_56065540 | 0.20 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr2_-_206950781 | 0.19 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr17_-_8301132 | 0.19 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chr8_-_145018905 | 0.19 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr16_-_1429010 | 0.19 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr11_-_117747327 | 0.18 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr14_+_71788096 | 0.17 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr4_-_84035905 | 0.17 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr5_-_176936844 | 0.17 |
ENST00000510380.1
ENST00000510898.1 ENST00000357198.4 |
DOK3
|
docking protein 3 |
| chr2_-_218843623 | 0.16 |
ENST00000413280.1
|
TNS1
|
tensin 1 |
| chr17_-_48278983 | 0.16 |
ENST00000225964.5
|
COL1A1
|
collagen, type I, alpha 1 |
| chr4_-_84035868 | 0.16 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr11_+_76092353 | 0.16 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr17_+_72427477 | 0.15 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_145780662 | 0.15 |
ENST00000423031.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr17_+_55333876 | 0.15 |
ENST00000284073.2
|
MSI2
|
musashi RNA-binding protein 2 |
| chr14_+_74815116 | 0.15 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chr3_-_123168551 | 0.15 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr15_-_88799948 | 0.15 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr19_+_47523058 | 0.15 |
ENST00000602212.1
ENST00000602189.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chr12_+_6881678 | 0.15 |
ENST00000441671.2
ENST00000203629.2 |
LAG3
|
lymphocyte-activation gene 3 |
| chr4_+_37892682 | 0.14 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr15_-_82338460 | 0.14 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr20_+_2083540 | 0.14 |
ENST00000400064.3
|
STK35
|
serine/threonine kinase 35 |
| chr11_-_130184470 | 0.14 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr16_-_18937726 | 0.14 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_+_167190066 | 0.14 |
ENST00000367866.2
ENST00000429375.2 ENST00000452019.1 ENST00000420254.3 ENST00000541643.3 |
POU2F1
|
POU class 2 homeobox 1 |
| chr12_+_57522692 | 0.14 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr19_+_50094866 | 0.14 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr17_-_36904437 | 0.14 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr17_-_47841485 | 0.14 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr17_+_7155343 | 0.13 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr8_+_123793633 | 0.13 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr20_+_61924532 | 0.13 |
ENST00000358894.6
ENST00000326996.6 ENST00000435874.1 |
COL20A1
|
collagen, type XX, alpha 1 |
| chr3_+_111393501 | 0.13 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr16_+_28505955 | 0.13 |
ENST00000564831.1
ENST00000328423.5 ENST00000431282.1 |
APOBR
|
apolipoprotein B receptor |
| chr8_+_145490549 | 0.13 |
ENST00000340695.2
|
SCXA
|
scleraxis homolog A (mouse) |
| chr12_+_57522439 | 0.13 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr19_-_12721616 | 0.13 |
ENST00000311437.6
|
ZNF490
|
zinc finger protein 490 |
| chr5_+_49962495 | 0.13 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr12_-_57914275 | 0.13 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr16_-_18937480 | 0.13 |
ENST00000532700.2
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr15_+_75640068 | 0.13 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr11_+_118307179 | 0.13 |
ENST00000534358.1
ENST00000531904.2 ENST00000389506.5 ENST00000354520.4 |
KMT2A
|
lysine (K)-specific methyltransferase 2A |
| chr15_-_37392086 | 0.13 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr2_-_145275211 | 0.12 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_29421900 | 0.12 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr3_+_63898275 | 0.12 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr19_+_34287751 | 0.12 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr19_-_14201776 | 0.12 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr3_+_110790590 | 0.12 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr1_+_6845578 | 0.11 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr22_-_27620603 | 0.11 |
ENST00000418271.1
ENST00000444114.1 |
RP5-1172A22.1
|
RP5-1172A22.1 |
| chr8_+_95907993 | 0.11 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr17_+_7211280 | 0.11 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr11_+_72525353 | 0.11 |
ENST00000321297.5
ENST00000534905.1 ENST00000540567.1 |
ATG16L2
|
autophagy related 16-like 2 (S. cerevisiae) |
| chr2_+_241564655 | 0.11 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35 |
| chr2_-_197458323 | 0.11 |
ENST00000452031.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_-_96076334 | 0.11 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr15_+_85144217 | 0.11 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr7_-_92463210 | 0.11 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr12_+_1738363 | 0.11 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr1_+_145516560 | 0.11 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr12_-_121972556 | 0.11 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr6_+_32936942 | 0.11 |
ENST00000496118.2
|
BRD2
|
bromodomain containing 2 |
| chr10_+_114709999 | 0.11 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_-_161147275 | 0.11 |
ENST00000319769.5
ENST00000367998.1 |
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr5_+_66254698 | 0.11 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_145018080 | 0.11 |
ENST00000354589.3
|
PLEC
|
plectin |
| chr6_-_152957944 | 0.11 |
ENST00000423061.1
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_155532484 | 0.11 |
ENST00000368346.3
ENST00000548830.1 |
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr8_+_27632083 | 0.10 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_+_145516252 | 0.10 |
ENST00000369306.3
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr6_-_32191834 | 0.10 |
ENST00000375023.3
|
NOTCH4
|
notch 4 |
| chr2_-_45166338 | 0.10 |
ENST00000437916.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr19_+_3178736 | 0.10 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr3_-_105587879 | 0.10 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr10_+_97803151 | 0.10 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr17_-_20370847 | 0.10 |
ENST00000423676.3
ENST00000324290.5 |
LGALS9B
|
lectin, galactoside-binding, soluble, 9B |
| chr10_+_105314881 | 0.10 |
ENST00000437579.1
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr17_+_40913264 | 0.10 |
ENST00000587142.1
ENST00000588576.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr4_+_153457404 | 0.10 |
ENST00000604157.1
ENST00000594836.1 |
MIR4453
|
microRNA 4453 |
| chrX_+_70316005 | 0.10 |
ENST00000374259.3
|
FOXO4
|
forkhead box O4 |
| chr22_-_41842781 | 0.10 |
ENST00000434408.1
|
TOB2
|
transducer of ERBB2, 2 |
| chr20_-_42816206 | 0.10 |
ENST00000372980.3
|
JPH2
|
junctophilin 2 |
| chr6_-_37665751 | 0.10 |
ENST00000297153.7
ENST00000434837.3 |
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr18_+_19321281 | 0.10 |
ENST00000261537.6
|
MIB1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr7_+_69064300 | 0.10 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr3_+_111393659 | 0.10 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr1_-_159915386 | 0.10 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr17_+_2496971 | 0.10 |
ENST00000397195.5
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
| chr16_+_77246337 | 0.09 |
ENST00000563157.1
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr3_+_184033551 | 0.09 |
ENST00000456033.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr9_+_2017063 | 0.09 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_371994 | 0.09 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr2_+_219264466 | 0.09 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr16_+_20911174 | 0.09 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr15_+_51973550 | 0.09 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr1_-_183560011 | 0.09 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr10_+_102672712 | 0.09 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr19_+_42788172 | 0.09 |
ENST00000160740.3
|
CIC
|
capicua transcriptional repressor |
| chr14_-_21994525 | 0.09 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr11_+_64002292 | 0.09 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr11_-_71751715 | 0.09 |
ENST00000535947.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr18_-_658244 | 0.09 |
ENST00000585033.1
ENST00000323813.3 |
C18orf56
|
chromosome 18 open reading frame 56 |
| chr4_-_41216473 | 0.09 |
ENST00000513140.1
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr3_+_14989076 | 0.09 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr5_-_81046841 | 0.09 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr6_+_33387868 | 0.09 |
ENST00000418600.2
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr17_-_42277203 | 0.09 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr3_+_187871659 | 0.09 |
ENST00000416784.1
ENST00000430340.1 ENST00000414139.1 ENST00000454789.1 |
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_86748863 | 0.09 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr14_+_32546274 | 0.09 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr2_-_75788424 | 0.09 |
ENST00000410071.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr11_+_46722368 | 0.09 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr6_-_32122106 | 0.09 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr1_+_211433275 | 0.08 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr19_-_36523529 | 0.08 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr11_+_82868185 | 0.08 |
ENST00000530304.1
ENST00000533018.1 |
PCF11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr7_-_100881109 | 0.08 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr3_+_148709310 | 0.08 |
ENST00000484197.1
ENST00000492285.2 ENST00000461191.1 |
GYG1
|
glycogenin 1 |
| chr16_+_69958887 | 0.08 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr17_-_42188598 | 0.08 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr19_-_38916839 | 0.08 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr15_-_83224682 | 0.08 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr13_-_30424821 | 0.08 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr6_-_151712673 | 0.08 |
ENST00000325144.4
|
ZBTB2
|
zinc finger and BTB domain containing 2 |
| chr1_-_24239844 | 0.08 |
ENST00000374472.4
|
CNR2
|
cannabinoid receptor 2 (macrophage) |
| chr7_-_100061869 | 0.08 |
ENST00000332375.3
|
C7orf61
|
chromosome 7 open reading frame 61 |
| chr17_-_58469329 | 0.08 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr7_-_100881041 | 0.08 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr15_-_37392703 | 0.08 |
ENST00000382766.2
ENST00000444725.1 |
MEIS2
|
Meis homeobox 2 |
| chr10_-_25305011 | 0.08 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr1_-_8585945 | 0.08 |
ENST00000377464.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr11_+_86748998 | 0.08 |
ENST00000525018.1
ENST00000355734.4 |
TMEM135
|
transmembrane protein 135 |
| chr11_-_119234876 | 0.08 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr17_+_72426891 | 0.08 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr3_+_88108381 | 0.08 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr9_+_34653861 | 0.08 |
ENST00000556792.1
ENST00000318041.9 ENST00000378817.4 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr2_-_26251481 | 0.08 |
ENST00000599234.1
|
AC013449.1
|
Uncharacterized protein |
| chr6_-_32160622 | 0.08 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr2_-_217236750 | 0.08 |
ENST00000273067.4
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
| chr9_+_130830451 | 0.08 |
ENST00000373068.2
ENST00000373069.5 |
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr12_-_15114603 | 0.08 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_-_53575120 | 0.08 |
ENST00000542115.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr19_+_50922187 | 0.08 |
ENST00000595883.1
ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr12_-_323248 | 0.08 |
ENST00000535347.1
ENST00000536824.1 |
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chrX_+_100878112 | 0.08 |
ENST00000491568.2
ENST00000479298.1 |
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr8_+_22250334 | 0.08 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr1_+_211432593 | 0.08 |
ENST00000367006.4
|
RCOR3
|
REST corepressor 3 |
| chrX_-_74743080 | 0.08 |
ENST00000373367.3
|
ZDHHC15
|
zinc finger, DHHC-type containing 15 |
| chr3_+_14989186 | 0.08 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr6_+_138483058 | 0.07 |
ENST00000251691.4
|
KIAA1244
|
KIAA1244 |
| chr12_+_7941989 | 0.07 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr10_-_103603568 | 0.07 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr1_+_14075903 | 0.07 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chrX_+_123095860 | 0.07 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr16_+_2802623 | 0.07 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr1_-_183559693 | 0.07 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr11_+_120195992 | 0.07 |
ENST00000314475.2
ENST00000529187.1 |
TMEM136
|
transmembrane protein 136 |
| chr12_+_57914742 | 0.07 |
ENST00000551351.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr1_+_200860122 | 0.07 |
ENST00000532631.1
ENST00000451872.2 |
C1orf106
|
chromosome 1 open reading frame 106 |
| chr6_+_111195973 | 0.07 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr3_+_8543393 | 0.07 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr19_-_6459746 | 0.07 |
ENST00000301454.4
ENST00000334510.5 |
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr5_+_158527485 | 0.07 |
ENST00000517335.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr1_+_221051699 | 0.07 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr2_+_203241531 | 0.07 |
ENST00000374580.4
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr3_+_184032919 | 0.07 |
ENST00000427845.1
ENST00000342981.4 ENST00000319274.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr22_-_37213045 | 0.07 |
ENST00000406910.2
ENST00000417718.2 |
PVALB
|
parvalbumin |
| chr1_+_16085263 | 0.07 |
ENST00000483633.2
ENST00000502739.1 ENST00000431771.2 |
FBLIM1
|
filamin binding LIM protein 1 |
| chrX_+_102883887 | 0.07 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr11_+_63449045 | 0.07 |
ENST00000354497.4
|
RTN3
|
reticulon 3 |
| chr4_-_121843985 | 0.07 |
ENST00000264808.3
ENST00000428209.2 ENST00000515109.1 ENST00000394435.2 |
PRDM5
|
PR domain containing 5 |
| chr19_-_40324255 | 0.07 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr20_-_42815733 | 0.07 |
ENST00000342272.3
|
JPH2
|
junctophilin 2 |
| chr7_+_143079000 | 0.07 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr17_-_46667628 | 0.07 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr3_-_113415441 | 0.07 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr7_+_76139833 | 0.07 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr17_-_40913275 | 0.07 |
ENST00000589716.1
ENST00000360166.3 |
RAMP2-AS1
|
RAMP2 antisense RNA 1 |
| chr2_-_61389240 | 0.07 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr3_+_8543533 | 0.07 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MZF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.2 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:2001113 | negative regulation of hepatocyte growth factor receptor signaling pathway(GO:1902203) regulation of cellular response to hepatocyte growth factor stimulus(GO:2001112) negative regulation of cellular response to hepatocyte growth factor stimulus(GO:2001113) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.1 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.1 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.2 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.0 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0046469 | platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:2000437 | egg activation(GO:0007343) monocyte extravasation(GO:0035696) regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.0 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.0 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.0 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.0 | 0.0 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.0 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.0 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.1 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.0 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |