Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
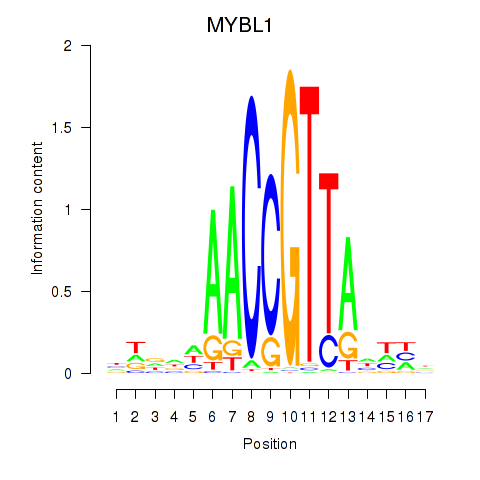
Results for MYBL1
Z-value: 0.86
Transcription factors associated with MYBL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL1
|
ENSG00000185697.12 | MYB proto-oncogene like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL1 | hg19_v2_chr8_-_67525524_67525543 | -0.69 | 1.3e-01 | Click! |
Activity profile of MYBL1 motif
Sorted Z-values of MYBL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_94982435 | 0.88 |
ENST00000511684.1
ENST00000380005.4 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr12_+_112451222 | 0.74 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr12_-_57081940 | 0.70 |
ENST00000436399.2
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr20_+_52824367 | 0.66 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr12_-_76478446 | 0.64 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr19_+_1495362 | 0.61 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr12_+_69753448 | 0.61 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr4_+_103790462 | 0.61 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr1_+_163291680 | 0.57 |
ENST00000450453.2
ENST00000524800.1 ENST00000442820.1 ENST00000367900.3 |
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr4_+_166248775 | 0.57 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr12_-_57082060 | 0.56 |
ENST00000448157.2
ENST00000414274.3 ENST00000262033.6 ENST00000456859.2 |
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr12_-_8693469 | 0.53 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr1_+_99127265 | 0.52 |
ENST00000306121.3
|
SNX7
|
sorting nexin 7 |
| chr7_-_34978980 | 0.51 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr8_-_25315905 | 0.51 |
ENST00000221200.4
|
KCTD9
|
potassium channel tetramerization domain containing 9 |
| chr3_-_182703688 | 0.51 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr19_-_6110555 | 0.50 |
ENST00000593241.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr2_-_87248975 | 0.50 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr12_-_76478686 | 0.49 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr16_-_56485257 | 0.49 |
ENST00000300291.5
|
NUDT21
|
nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
| chr19_+_36239576 | 0.47 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr7_+_64254766 | 0.45 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chrM_+_4431 | 0.44 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr17_+_48243352 | 0.43 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr7_+_7222233 | 0.43 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr3_-_148804275 | 0.42 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr3_-_100120223 | 0.42 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr6_+_142468361 | 0.42 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr4_+_76649753 | 0.41 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr5_+_61708582 | 0.41 |
ENST00000325324.6
|
IPO11
|
importin 11 |
| chr10_-_35379524 | 0.41 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr7_-_124569991 | 0.41 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr19_+_40502938 | 0.40 |
ENST00000599504.1
ENST00000596894.1 ENST00000601138.1 ENST00000600094.1 ENST00000347077.4 |
ZNF546
|
zinc finger protein 546 |
| chr1_+_97187318 | 0.40 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr1_+_173991633 | 0.40 |
ENST00000424181.1
|
RP11-160H22.3
|
RP11-160H22.3 |
| chr3_+_49236910 | 0.39 |
ENST00000452691.2
ENST00000366429.2 |
CCDC36
|
coiled-coil domain containing 36 |
| chr11_+_33037652 | 0.39 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr4_+_76649797 | 0.39 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr4_-_120988229 | 0.38 |
ENST00000296509.6
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr1_+_99127225 | 0.38 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr13_+_32889605 | 0.37 |
ENST00000380152.3
ENST00000544455.1 ENST00000530893.2 |
BRCA2
|
breast cancer 2, early onset |
| chr7_-_151217166 | 0.36 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr14_-_58893832 | 0.36 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr10_+_60145155 | 0.34 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr1_+_163291732 | 0.34 |
ENST00000271452.3
|
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr7_-_124569864 | 0.34 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr1_-_197115818 | 0.34 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr14_-_58894223 | 0.33 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr1_+_186344945 | 0.33 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr16_-_25122735 | 0.33 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr2_-_47403642 | 0.33 |
ENST00000456319.1
ENST00000409563.1 ENST00000272298.7 |
CALM2
|
calmodulin 2 (phosphorylase kinase, delta) |
| chr21_-_43816152 | 0.33 |
ENST00000433957.2
ENST00000398397.3 |
TMPRSS3
|
transmembrane protease, serine 3 |
| chr1_-_193074504 | 0.33 |
ENST00000367439.3
|
GLRX2
|
glutaredoxin 2 |
| chr3_+_73045936 | 0.33 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr1_-_68962805 | 0.32 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chrM_+_9207 | 0.32 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr15_-_55489097 | 0.32 |
ENST00000260443.4
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr3_-_134204815 | 0.32 |
ENST00000514612.1
ENST00000510994.1 ENST00000354910.5 |
ANAPC13
|
anaphase promoting complex subunit 13 |
| chr14_-_58893876 | 0.31 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr12_-_66563728 | 0.31 |
ENST00000286424.7
ENST00000556010.1 ENST00000398033.4 |
TMBIM4
|
transmembrane BAX inhibitor motif containing 4 |
| chr14_+_45366472 | 0.31 |
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr3_-_160117301 | 0.31 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr11_+_18433840 | 0.30 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr8_-_95565673 | 0.30 |
ENST00000437199.1
ENST00000297591.5 ENST00000421249.2 |
KIAA1429
|
KIAA1429 |
| chr8_-_119634141 | 0.29 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr8_+_101162812 | 0.29 |
ENST00000353107.3
ENST00000522439.1 |
POLR2K
|
polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa |
| chr3_-_10052869 | 0.29 |
ENST00000454232.1
|
AC022007.5
|
AC022007.5 |
| chr8_+_25316489 | 0.29 |
ENST00000330560.3
|
CDCA2
|
cell division cycle associated 2 |
| chr18_+_52385068 | 0.28 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr2_+_196521845 | 0.28 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr8_+_25316707 | 0.28 |
ENST00000380665.3
|
CDCA2
|
cell division cycle associated 2 |
| chr13_-_49987885 | 0.28 |
ENST00000409082.1
|
CAB39L
|
calcium binding protein 39-like |
| chr1_+_172502336 | 0.28 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr3_-_192445289 | 0.27 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr3_+_112709804 | 0.27 |
ENST00000383677.3
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr12_+_104697504 | 0.27 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr1_+_162467595 | 0.27 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr3_+_97483572 | 0.27 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr5_+_94982558 | 0.27 |
ENST00000311364.4
ENST00000458310.1 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr14_+_57857262 | 0.26 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr2_-_55647057 | 0.26 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr14_-_58894332 | 0.26 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr2_-_191115229 | 0.26 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr12_+_107349497 | 0.26 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr6_+_142468383 | 0.26 |
ENST00000367621.1
ENST00000452973.2 |
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr3_+_32433154 | 0.26 |
ENST00000334983.5
ENST00000349718.4 |
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr14_+_45366518 | 0.26 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr20_+_5986727 | 0.26 |
ENST00000378863.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_+_172502244 | 0.25 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr6_+_168434678 | 0.25 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr2_-_86564740 | 0.25 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr10_-_91174215 | 0.24 |
ENST00000371837.1
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr3_+_12598563 | 0.24 |
ENST00000411987.1
ENST00000448482.1 |
MKRN2
|
makorin ring finger protein 2 |
| chr8_+_38244638 | 0.24 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr11_-_92930556 | 0.24 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr2_+_208576259 | 0.24 |
ENST00000392209.3
|
CCNYL1
|
cyclin Y-like 1 |
| chr6_+_122793058 | 0.24 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr19_-_55672037 | 0.24 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr18_+_20513782 | 0.24 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr15_-_63449663 | 0.24 |
ENST00000439025.1
|
RPS27L
|
ribosomal protein S27-like |
| chr2_-_55646957 | 0.23 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr15_+_38746307 | 0.23 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr8_-_91997427 | 0.23 |
ENST00000517562.2
|
RP11-122A3.2
|
chromosome 8 open reading frame 88 |
| chr15_+_66797455 | 0.23 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr16_+_67360856 | 0.23 |
ENST00000568804.2
|
LRRC36
|
leucine rich repeat containing 36 |
| chr7_+_39605966 | 0.22 |
ENST00000223273.2
ENST00000448268.1 ENST00000432096.2 |
YAE1D1
|
Yae1 domain containing 1 |
| chr15_+_78730531 | 0.22 |
ENST00000258886.8
|
IREB2
|
iron-responsive element binding protein 2 |
| chr8_+_124084899 | 0.22 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr4_+_17812525 | 0.22 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr8_+_104426942 | 0.22 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr9_+_4679555 | 0.22 |
ENST00000381858.1
ENST00000381854.3 |
CDC37L1
|
cell division cycle 37-like 1 |
| chr6_+_88182643 | 0.22 |
ENST00000369556.3
ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr15_+_66797627 | 0.22 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr15_-_96590126 | 0.21 |
ENST00000561051.1
|
RP11-4G2.1
|
RP11-4G2.1 |
| chr2_-_201936302 | 0.21 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr3_-_176915215 | 0.21 |
ENST00000457928.2
ENST00000422442.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr5_-_146833803 | 0.21 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr17_-_30669138 | 0.21 |
ENST00000225805.4
ENST00000577809.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr14_-_50101931 | 0.21 |
ENST00000298292.8
ENST00000406043.3 |
DNAAF2
|
dynein, axonemal, assembly factor 2 |
| chr1_-_63782888 | 0.21 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr21_-_16254231 | 0.20 |
ENST00000412426.1
ENST00000418954.1 |
AF127936.7
|
AF127936.7 |
| chr3_+_97483366 | 0.20 |
ENST00000463745.1
ENST00000462412.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr5_-_10249990 | 0.20 |
ENST00000511437.1
ENST00000280330.8 ENST00000510047.1 |
FAM173B
|
family with sequence similarity 173, member B |
| chr1_+_236686875 | 0.20 |
ENST00000366584.4
|
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chrX_-_71458802 | 0.20 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr7_+_23221438 | 0.20 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr5_-_74162605 | 0.20 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr2_+_69969106 | 0.19 |
ENST00000409920.1
ENST00000394295.4 ENST00000536030.1 |
ANXA4
|
annexin A4 |
| chr2_+_208576355 | 0.19 |
ENST00000420822.1
ENST00000295414.3 ENST00000339882.5 |
CCNYL1
|
cyclin Y-like 1 |
| chr2_-_68547019 | 0.19 |
ENST00000409862.1
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr6_-_28554977 | 0.19 |
ENST00000452236.2
|
SCAND3
|
SCAN domain containing 3 |
| chr11_+_29181503 | 0.19 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr14_+_60558627 | 0.19 |
ENST00000317623.4
ENST00000391611.2 ENST00000406854.1 ENST00000406949.1 |
PCNXL4
|
pecanex-like 4 (Drosophila) |
| chr11_+_57365150 | 0.19 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr19_-_52551814 | 0.19 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr14_+_58894103 | 0.19 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr10_+_91174314 | 0.19 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr5_-_75919217 | 0.18 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr3_+_134204551 | 0.18 |
ENST00000332047.5
ENST00000354446.3 |
CEP63
|
centrosomal protein 63kDa |
| chr4_-_4544061 | 0.18 |
ENST00000507908.1
|
STX18
|
syntaxin 18 |
| chr16_-_20753114 | 0.18 |
ENST00000396083.2
|
THUMPD1
|
THUMP domain containing 1 |
| chr19_-_36643329 | 0.18 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr7_+_12726474 | 0.18 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr2_-_86564776 | 0.18 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr18_+_3247413 | 0.18 |
ENST00000579226.1
ENST00000217652.3 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr11_-_3013497 | 0.18 |
ENST00000448187.1
ENST00000532325.2 ENST00000399614.2 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr3_-_3221358 | 0.17 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr8_+_124780672 | 0.17 |
ENST00000521166.1
ENST00000334705.7 |
FAM91A1
|
family with sequence similarity 91, member A1 |
| chr4_-_147443043 | 0.17 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr19_+_44764031 | 0.17 |
ENST00000592581.1
ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233
|
zinc finger protein 233 |
| chr4_+_79567362 | 0.17 |
ENST00000512322.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr6_+_27782788 | 0.17 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr2_-_178483694 | 0.17 |
ENST00000355689.5
|
TTC30A
|
tetratricopeptide repeat domain 30A |
| chr8_-_104427289 | 0.17 |
ENST00000543107.1
|
SLC25A32
|
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr8_+_104384616 | 0.17 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr8_-_96281419 | 0.17 |
ENST00000286688.5
|
C8orf37
|
chromosome 8 open reading frame 37 |
| chr2_+_29353520 | 0.16 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr4_+_119200215 | 0.16 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr2_+_95963052 | 0.16 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr8_-_82598511 | 0.16 |
ENST00000449740.2
ENST00000311489.4 ENST00000521360.1 ENST00000519964.1 ENST00000518202.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr2_+_201936458 | 0.16 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr18_-_44497308 | 0.16 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr8_-_133687813 | 0.16 |
ENST00000250173.1
ENST00000519595.1 |
LRRC6
|
leucine rich repeat containing 6 |
| chr2_-_44223138 | 0.16 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr3_-_10052763 | 0.15 |
ENST00000383808.2
ENST00000426698.1 ENST00000470827.2 |
AC022007.5
EMC3
|
AC022007.5 ER membrane protein complex subunit 3 |
| chr4_+_185734773 | 0.15 |
ENST00000508020.1
|
RP11-701P16.2
|
Uncharacterized protein |
| chr22_+_22723969 | 0.15 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr3_-_52739670 | 0.15 |
ENST00000497953.1
|
GLT8D1
|
glycosyltransferase 8 domain containing 1 |
| chr3_+_134205000 | 0.15 |
ENST00000512894.1
ENST00000513612.2 ENST00000606977.1 |
CEP63
|
centrosomal protein 63kDa |
| chr8_+_104310661 | 0.15 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr5_-_95018660 | 0.14 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr12_+_133614062 | 0.14 |
ENST00000540031.1
ENST00000536123.1 |
ZNF84
|
zinc finger protein 84 |
| chr3_-_10052849 | 0.14 |
ENST00000437616.1
ENST00000429065.2 |
AC022007.5
|
AC022007.5 |
| chr11_-_104840093 | 0.14 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr3_-_142166904 | 0.14 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr6_+_36562132 | 0.14 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chr22_-_28315115 | 0.14 |
ENST00000455418.3
ENST00000436663.1 ENST00000320996.10 ENST00000335272.5 |
PITPNB
|
phosphatidylinositol transfer protein, beta |
| chr8_-_100905850 | 0.14 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr4_-_140098339 | 0.14 |
ENST00000394235.2
|
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr8_+_92082424 | 0.14 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chr20_-_1309809 | 0.14 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr2_+_9563697 | 0.14 |
ENST00000238112.3
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr4_+_174818390 | 0.14 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr12_-_104359475 | 0.14 |
ENST00000553183.1
|
C12orf73
|
chromosome 12 open reading frame 73 |
| chr2_-_40006357 | 0.13 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr11_+_10326918 | 0.13 |
ENST00000528544.1
|
ADM
|
adrenomedullin |
| chr7_-_139763521 | 0.13 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr2_-_182521699 | 0.13 |
ENST00000374969.2
ENST00000339098.5 ENST00000374970.2 |
CERKL
|
ceramide kinase-like |
| chr7_-_151217001 | 0.13 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr12_+_25055243 | 0.13 |
ENST00000599478.1
|
AC026310.1
|
Protein 101060047 |
| chr10_+_104613980 | 0.13 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr1_+_210406121 | 0.13 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr18_-_32924372 | 0.13 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr17_-_48133054 | 0.13 |
ENST00000499842.1
|
RP11-1094H24.4
|
RP11-1094H24.4 |
| chr11_+_20409070 | 0.13 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr14_+_58894141 | 0.13 |
ENST00000423743.3
|
KIAA0586
|
KIAA0586 |
| chr19_-_13030071 | 0.13 |
ENST00000293695.7
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chr20_+_18548055 | 0.12 |
ENST00000435844.1
ENST00000411646.1 ENST00000608034.1 |
LINC00493
|
long intergenic non-protein coding RNA 493 |
| chr21_-_15755446 | 0.12 |
ENST00000544452.1
ENST00000285667.3 |
HSPA13
|
heat shock protein 70kDa family, member 13 |
| chr1_-_84464780 | 0.12 |
ENST00000260505.8
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.4 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.1 | 1.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.5 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 0.3 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.4 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 1.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.2 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.5 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.7 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.4 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:1903947 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.3 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.3 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:1903381 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.2 | GO:0015827 | L-alanine transport(GO:0015808) tryptophan transport(GO:0015827) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.5 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.0 | 0.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 1.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 0.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.3 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.3 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.1 | 2.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.2 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 2.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.0 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |