Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
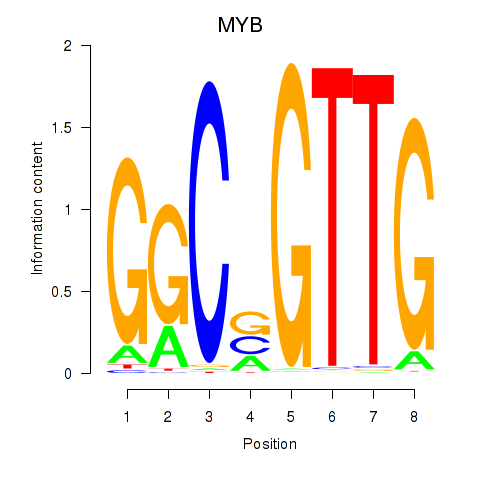
Results for MYB
Z-value: 1.15
Transcription factors associated with MYB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYB
|
ENSG00000118513.14 | MYB proto-oncogene, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYB | hg19_v2_chr6_+_135502466_135502489 | -0.15 | 7.8e-01 | Click! |
Activity profile of MYB motif
Sorted Z-values of MYB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_100967533 | 0.78 |
ENST00000550295.1
|
GAS2L3
|
growth arrest-specific 2 like 3 |
| chr11_-_130184555 | 0.70 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr5_+_98264867 | 0.67 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr3_-_88108192 | 0.67 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr10_-_98347063 | 0.62 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr2_-_61244550 | 0.59 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr11_+_130184888 | 0.57 |
ENST00000602376.1
ENST00000532116.3 ENST00000602310.1 |
RP11-121M22.1
|
RP11-121M22.1 |
| chr9_-_4679419 | 0.53 |
ENST00000609131.1
ENST00000607997.1 |
RP11-6J24.6
|
RP11-6J24.6 |
| chr7_-_127032363 | 0.52 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr4_+_44019074 | 0.49 |
ENST00000512678.1
|
RP11-328N19.1
|
RP11-328N19.1 |
| chr4_-_101111615 | 0.48 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr13_-_52026730 | 0.46 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr9_+_4679555 | 0.45 |
ENST00000381858.1
ENST00000381854.3 |
CDC37L1
|
cell division cycle 37-like 1 |
| chr1_-_100598444 | 0.45 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr14_+_61449197 | 0.43 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr2_-_61245363 | 0.42 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr2_-_176866978 | 0.41 |
ENST00000392540.2
ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715
|
KIAA1715 |
| chr17_-_33288419 | 0.41 |
ENST00000421975.3
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2) |
| chr3_-_178865747 | 0.39 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr7_+_8008418 | 0.38 |
ENST00000223145.5
|
GLCCI1
|
glucocorticoid induced transcript 1 |
| chr13_-_103426081 | 0.38 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr11_-_130184470 | 0.38 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr11_+_65265141 | 0.37 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr6_+_13925318 | 0.36 |
ENST00000423553.2
ENST00000537388.1 |
RNF182
|
ring finger protein 182 |
| chr8_-_101964265 | 0.34 |
ENST00000395958.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr7_-_151217166 | 0.34 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr12_-_57081940 | 0.34 |
ENST00000436399.2
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr1_-_68962744 | 0.33 |
ENST00000525124.1
|
DEPDC1
|
DEP domain containing 1 |
| chr10_-_105238997 | 0.33 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chr8_+_67782984 | 0.32 |
ENST00000396592.3
ENST00000422365.2 ENST00000492775.1 |
MCMDC2
|
minichromosome maintenance domain containing 2 |
| chr1_-_85156090 | 0.32 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr2_+_196521903 | 0.32 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_+_109234907 | 0.32 |
ENST00000370025.4
ENST00000370022.5 ENST00000370021.1 |
PRPF38B
|
pre-mRNA processing factor 38B |
| chr1_+_178694408 | 0.32 |
ENST00000324778.5
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr5_+_56469843 | 0.32 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chrX_+_23682379 | 0.31 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr17_-_33288522 | 0.31 |
ENST00000314144.5
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2) |
| chr3_+_88108381 | 0.31 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr8_+_17780346 | 0.31 |
ENST00000325083.8
|
PCM1
|
pericentriolar material 1 |
| chr4_-_156298087 | 0.31 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr8_-_71519889 | 0.30 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr7_-_151107767 | 0.30 |
ENST00000477459.1
|
WDR86
|
WD repeat domain 86 |
| chr1_+_60280458 | 0.30 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr12_+_31079652 | 0.30 |
ENST00000546076.1
ENST00000535215.1 ENST00000544427.1 ENST00000261177.9 |
TSPAN11
|
tetraspanin 11 |
| chr4_-_141348763 | 0.30 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr13_-_103426112 | 0.30 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr1_-_85156216 | 0.29 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr11_+_65266507 | 0.29 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr9_-_36400920 | 0.28 |
ENST00000357058.3
ENST00000350199.4 |
RNF38
|
ring finger protein 38 |
| chr12_-_57082060 | 0.27 |
ENST00000448157.2
ENST00000414274.3 ENST00000262033.6 ENST00000456859.2 |
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr16_-_48419361 | 0.27 |
ENST00000394725.2
|
SIAH1
|
siah E3 ubiquitin protein ligase 1 |
| chr7_+_65338312 | 0.27 |
ENST00000434382.2
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr10_-_64028466 | 0.27 |
ENST00000395265.1
ENST00000373789.3 ENST00000395260.3 |
RTKN2
|
rhotekin 2 |
| chr3_-_160117301 | 0.26 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr1_+_172502336 | 0.26 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr11_+_34073195 | 0.26 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr9_+_108320392 | 0.26 |
ENST00000602661.1
ENST00000223528.2 ENST00000448551.2 ENST00000540160.1 |
FKTN
|
fukutin |
| chr14_-_36277857 | 0.25 |
ENST00000553892.1
ENST00000382366.3 |
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr2_+_233385173 | 0.25 |
ENST00000449534.2
|
PRSS56
|
protease, serine, 56 |
| chr18_+_34409069 | 0.25 |
ENST00000543923.1
ENST00000280020.5 ENST00000435985.2 ENST00000592521.1 ENST00000587139.1 |
KIAA1328
|
KIAA1328 |
| chr6_-_149969829 | 0.25 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr3_+_197476621 | 0.25 |
ENST00000241502.4
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr6_-_100016492 | 0.25 |
ENST00000369217.4
ENST00000369220.4 ENST00000482541.2 |
CCNC
|
cyclin C |
| chr1_-_68962805 | 0.25 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr10_-_46641003 | 0.24 |
ENST00000395721.2
ENST00000374218.2 ENST00000395725.3 ENST00000374346.3 ENST00000417004.1 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr2_+_201390843 | 0.24 |
ENST00000357799.4
ENST00000409203.3 |
SGOL2
|
shugoshin-like 2 (S. pombe) |
| chr10_-_124713842 | 0.24 |
ENST00000481909.1
|
C10orf88
|
chromosome 10 open reading frame 88 |
| chr19_-_49016847 | 0.24 |
ENST00000598924.1
|
CTC-273B12.10
|
CTC-273B12.10 |
| chr10_-_98346801 | 0.24 |
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr9_+_79792410 | 0.24 |
ENST00000357409.5
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr8_-_53626974 | 0.24 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr12_-_66563786 | 0.24 |
ENST00000542724.1
|
TMBIM4
|
transmembrane BAX inhibitor motif containing 4 |
| chr17_+_40950797 | 0.24 |
ENST00000588408.1
ENST00000585355.1 |
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr3_-_160117035 | 0.24 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr15_-_72410455 | 0.24 |
ENST00000569314.1
|
MYO9A
|
myosin IXA |
| chr4_-_141348789 | 0.24 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr21_-_35014027 | 0.24 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr10_+_116853201 | 0.23 |
ENST00000527407.1
|
ATRNL1
|
attractin-like 1 |
| chr3_-_182698381 | 0.23 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr8_-_101964231 | 0.23 |
ENST00000521309.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_-_49198095 | 0.23 |
ENST00000505279.1
|
SPAG9
|
sperm associated antigen 9 |
| chr17_-_45056606 | 0.23 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr13_-_108867846 | 0.23 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr16_-_15982440 | 0.22 |
ENST00000575938.1
ENST00000573396.1 ENST00000573968.1 ENST00000575744.1 ENST00000573429.1 ENST00000255759.6 ENST00000575073.1 |
FOPNL
|
FGFR1OP N-terminal like |
| chr16_+_19535133 | 0.22 |
ENST00000396212.2
ENST00000381396.5 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr18_+_56338618 | 0.22 |
ENST00000348428.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr1_-_68962782 | 0.22 |
ENST00000456315.2
|
DEPDC1
|
DEP domain containing 1 |
| chr5_+_56469939 | 0.22 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr2_-_38830090 | 0.22 |
ENST00000449105.3
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr9_+_79792269 | 0.22 |
ENST00000376634.4
ENST00000376636.3 ENST00000360280.3 |
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr3_-_24536453 | 0.22 |
ENST00000453729.2
ENST00000413780.1 |
THRB
|
thyroid hormone receptor, beta |
| chr1_+_172502244 | 0.22 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr1_+_100598742 | 0.22 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr8_+_96145974 | 0.21 |
ENST00000315367.3
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr15_-_35280426 | 0.21 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr2_+_196521845 | 0.21 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr17_-_17109579 | 0.21 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr14_+_71788096 | 0.21 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr4_-_146019335 | 0.21 |
ENST00000451299.2
ENST00000507656.1 ENST00000309439.5 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr6_-_41040049 | 0.21 |
ENST00000471367.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr8_+_17780483 | 0.21 |
ENST00000517730.1
ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1
|
pericentriolar material 1 |
| chr2_-_47403642 | 0.21 |
ENST00000456319.1
ENST00000409563.1 ENST00000272298.7 |
CALM2
|
calmodulin 2 (phosphorylase kinase, delta) |
| chr16_+_67063262 | 0.21 |
ENST00000565389.1
|
CBFB
|
core-binding factor, beta subunit |
| chr16_+_50059182 | 0.21 |
ENST00000562576.1
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr16_-_67260901 | 0.21 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr2_-_38829990 | 0.21 |
ENST00000409328.1
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr16_-_3285144 | 0.21 |
ENST00000431561.3
ENST00000396870.4 |
ZNF200
|
zinc finger protein 200 |
| chr1_+_225117350 | 0.20 |
ENST00000413949.2
ENST00000430092.1 ENST00000366850.3 ENST00000400952.3 ENST00000366849.1 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr3_-_160116995 | 0.20 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr18_-_268019 | 0.20 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chr1_-_200379180 | 0.20 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chrX_+_133507327 | 0.20 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr1_-_101360331 | 0.20 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr15_+_59499031 | 0.20 |
ENST00000307144.4
|
LDHAL6B
|
lactate dehydrogenase A-like 6B |
| chr12_+_100967420 | 0.20 |
ENST00000266754.5
ENST00000547754.1 |
GAS2L3
|
growth arrest-specific 2 like 3 |
| chr14_-_92572894 | 0.20 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chrX_+_49969405 | 0.20 |
ENST00000376042.1
|
CCNB3
|
cyclin B3 |
| chr17_-_16395328 | 0.20 |
ENST00000470794.1
|
FAM211A
|
family with sequence similarity 211, member A |
| chr3_+_122920847 | 0.20 |
ENST00000466519.1
ENST00000480631.1 ENST00000491366.1 ENST00000487572.1 |
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae) |
| chr16_-_56485257 | 0.20 |
ENST00000300291.5
|
NUDT21
|
nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
| chr5_-_114961858 | 0.20 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr15_+_72410629 | 0.19 |
ENST00000340912.4
ENST00000544171.1 |
SENP8
|
SUMO/sentrin specific peptidase family member 8 |
| chr6_+_64282447 | 0.19 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr1_-_229406746 | 0.19 |
ENST00000429227.1
ENST00000436334.1 |
RP5-1061H20.4
|
RP5-1061H20.4 |
| chr21_-_15755446 | 0.19 |
ENST00000544452.1
ENST00000285667.3 |
HSPA13
|
heat shock protein 70kDa family, member 13 |
| chrX_+_133507283 | 0.19 |
ENST00000370803.3
|
PHF6
|
PHD finger protein 6 |
| chr3_+_31574189 | 0.19 |
ENST00000295770.2
|
STT3B
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr17_+_42733803 | 0.19 |
ENST00000409122.2
|
C17orf104
|
chromosome 17 open reading frame 104 |
| chr17_+_74733744 | 0.19 |
ENST00000586689.1
ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr6_+_87865262 | 0.19 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr8_+_86019382 | 0.19 |
ENST00000360375.3
|
LRRCC1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr9_+_74764340 | 0.19 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr13_-_52027134 | 0.19 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr15_-_72410350 | 0.19 |
ENST00000356056.5
ENST00000424560.1 ENST00000444904.1 |
MYO9A
|
myosin IXA |
| chr2_-_153574480 | 0.19 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr18_-_6929797 | 0.19 |
ENST00000581725.1
ENST00000583316.1 |
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr10_+_32735177 | 0.19 |
ENST00000545067.1
|
CCDC7
|
coiled-coil domain containing 7 |
| chr4_-_125633876 | 0.19 |
ENST00000504087.1
ENST00000515641.1 |
ANKRD50
|
ankyrin repeat domain 50 |
| chr1_+_52195480 | 0.19 |
ENST00000531828.1
ENST00000361556.5 ENST00000481937.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr11_+_34073757 | 0.19 |
ENST00000532820.1
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr16_+_67918708 | 0.19 |
ENST00000339176.3
ENST00000576758.1 |
NRN1L
|
neuritin 1-like |
| chr9_+_15553055 | 0.18 |
ENST00000380701.3
|
CCDC171
|
coiled-coil domain containing 171 |
| chr3_-_123680246 | 0.18 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr5_+_56469775 | 0.18 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr1_+_52195542 | 0.18 |
ENST00000462759.1
ENST00000486942.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr1_-_26232522 | 0.18 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr14_+_96968802 | 0.18 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr18_-_47013586 | 0.18 |
ENST00000318240.3
ENST00000579820.1 |
C18orf32
|
chromosome 18 open reading frame 32 |
| chr6_+_13925170 | 0.18 |
ENST00000471906.1
|
RNF182
|
ring finger protein 182 |
| chr2_+_29033682 | 0.18 |
ENST00000379579.4
ENST00000334056.5 ENST00000449210.1 |
SPDYA
|
speedy/RINGO cell cycle regulator family member A |
| chr13_+_60971080 | 0.18 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr10_-_21463116 | 0.18 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr3_+_197477038 | 0.18 |
ENST00000426031.1
ENST00000424384.2 |
FYTTD1
|
forty-two-three domain containing 1 |
| chr16_-_46865047 | 0.18 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr3_+_160117087 | 0.18 |
ENST00000357388.3
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr13_-_41837620 | 0.18 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr6_-_41040195 | 0.18 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr16_+_16481306 | 0.18 |
ENST00000422673.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr3_-_47205066 | 0.18 |
ENST00000412450.1
|
SETD2
|
SET domain containing 2 |
| chr7_-_105752651 | 0.17 |
ENST00000470347.1
ENST00000455385.2 |
SYPL1
|
synaptophysin-like 1 |
| chr2_-_9770706 | 0.17 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr2_-_39347524 | 0.17 |
ENST00000395038.2
ENST00000402219.2 |
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr5_+_68665608 | 0.17 |
ENST00000509734.1
ENST00000354868.5 ENST00000521422.1 ENST00000354312.3 ENST00000345306.6 |
RAD17
|
RAD17 homolog (S. pombe) |
| chr14_+_74111578 | 0.17 |
ENST00000554113.1
ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1
|
dynein, axonemal, light chain 1 |
| chr15_-_43212836 | 0.17 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr9_-_36401155 | 0.17 |
ENST00000377885.2
|
RNF38
|
ring finger protein 38 |
| chr16_-_71598823 | 0.17 |
ENST00000566202.1
|
ZNF19
|
zinc finger protein 19 |
| chr1_-_114355083 | 0.17 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr1_+_174128639 | 0.17 |
ENST00000251507.4
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr10_+_92980517 | 0.17 |
ENST00000336126.5
|
PCGF5
|
polycomb group ring finger 5 |
| chr8_+_124084899 | 0.17 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr17_-_7197881 | 0.17 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chr16_+_69985644 | 0.17 |
ENST00000561889.1
|
CLEC18A
|
C-type lectin domain family 18, member A |
| chr19_+_38826477 | 0.17 |
ENST00000409410.2
ENST00000215069.4 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr2_-_69870747 | 0.17 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr10_-_46640660 | 0.17 |
ENST00000395727.2
ENST00000509900.1 ENST00000503851.1 ENST00000506080.1 ENST00000513266.1 ENST00000502705.1 ENST00000505814.1 ENST00000509774.1 ENST00000511769.1 ENST00000509599.1 ENST00000513156.1 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr5_+_74633036 | 0.17 |
ENST00000343975.5
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr16_+_50059125 | 0.17 |
ENST00000427478.2
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr2_+_46926326 | 0.17 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr1_+_41707996 | 0.17 |
ENST00000425554.1
|
RP11-399E6.1
|
RP11-399E6.1 |
| chr2_-_38830160 | 0.17 |
ENST00000409636.1
ENST00000608859.1 ENST00000358367.4 |
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr13_-_26795840 | 0.17 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr18_+_43684298 | 0.17 |
ENST00000282058.6
|
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr17_-_58499766 | 0.16 |
ENST00000588898.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr6_+_167412665 | 0.16 |
ENST00000366847.4
|
FGFR1OP
|
FGFR1 oncogene partner |
| chr10_-_92617437 | 0.16 |
ENST00000336152.3
ENST00000277874.6 ENST00000371719.2 |
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
| chr6_+_71123107 | 0.16 |
ENST00000370479.3
ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A
|
family with sequence similarity 135, member A |
| chr14_+_54863682 | 0.16 |
ENST00000543789.2
ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr10_+_91461337 | 0.16 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr2_+_242289502 | 0.16 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr1_+_2066387 | 0.16 |
ENST00000497183.1
|
PRKCZ
|
protein kinase C, zeta |
| chr5_-_149669612 | 0.16 |
ENST00000510347.1
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr6_+_26521948 | 0.16 |
ENST00000411553.1
|
HCG11
|
HLA complex group 11 (non-protein coding) |
| chr10_-_101945771 | 0.16 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr14_+_79745682 | 0.16 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr2_-_174828892 | 0.16 |
ENST00000418194.2
|
SP3
|
Sp3 transcription factor |
| chr2_+_196521458 | 0.16 |
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr17_+_47439733 | 0.16 |
ENST00000507337.1
|
RP11-1079K10.3
|
RP11-1079K10.3 |
| chr2_-_44588893 | 0.16 |
ENST00000409272.1
ENST00000410081.1 ENST00000541738.1 |
PREPL
|
prolyl endopeptidase-like |
| chr14_+_54863667 | 0.16 |
ENST00000335183.6
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.3 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 1.0 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.3 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 0.6 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.2 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.4 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.2 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.1 | 0.2 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.2 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.2 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0061570 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.2 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.2 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.2 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.7 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.7 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) female pronucleus assembly(GO:0035038) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0036102 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) choline metabolic process(GO:0019695) |
| 0.0 | 0.2 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 1.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0002254 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0014735 | regulation of muscle atrophy(GO:0014735) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:1901490 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.8 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.2 | GO:0007099 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.5 | GO:0032967 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.5 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.4 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.2 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.8 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.6 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.1 | 0.3 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) choline binding(GO:0033265) |
| 0.1 | 0.4 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.2 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 0.3 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.0 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |