Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for MSX1
Z-value: 0.59
Transcription factors associated with MSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX1
|
ENSG00000163132.6 | msh homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX1 | hg19_v2_chr4_+_4861385_4861398 | -0.11 | 8.4e-01 | Click! |
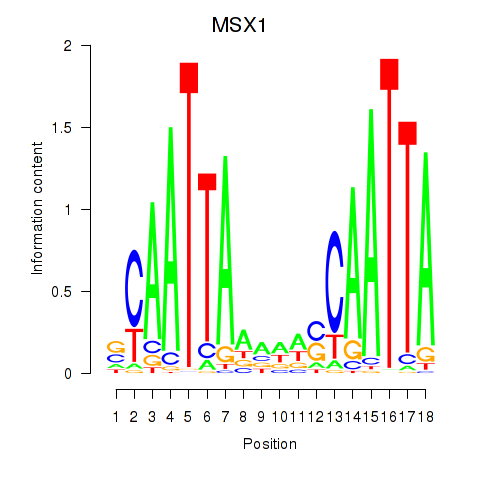
Activity profile of MSX1 motif
Sorted Z-values of MSX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_115498761 | 0.66 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr7_+_99425633 | 0.45 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr2_+_7073174 | 0.36 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr14_-_92247032 | 0.34 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr5_+_121465234 | 0.34 |
ENST00000504912.1
ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr7_-_105332084 | 0.30 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr8_-_25281747 | 0.30 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr4_-_100140331 | 0.25 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr19_+_3762645 | 0.25 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chrX_+_23928500 | 0.25 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr13_-_38172863 | 0.24 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr4_+_169013666 | 0.24 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr1_-_20503917 | 0.23 |
ENST00000429261.2
|
PLA2G2C
|
phospholipase A2, group IIC |
| chr2_+_175352114 | 0.22 |
ENST00000444196.1
ENST00000417038.1 ENST00000606406.1 |
AC010894.3
|
AC010894.3 |
| chr16_+_53412368 | 0.21 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr17_-_47786375 | 0.20 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr5_-_119669160 | 0.20 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr22_-_32767017 | 0.19 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr15_+_77713222 | 0.19 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr21_+_33671264 | 0.19 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr13_-_81801115 | 0.18 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr6_+_134758827 | 0.18 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr12_+_27623565 | 0.18 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr6_+_131958436 | 0.17 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr12_+_48592134 | 0.17 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr14_+_52327350 | 0.17 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr6_-_31830655 | 0.17 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr10_-_61899124 | 0.16 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr5_-_75919253 | 0.16 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr14_+_20215587 | 0.15 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr3_+_187957646 | 0.15 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr14_-_101295407 | 0.14 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr21_+_43619796 | 0.14 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr8_+_97597148 | 0.14 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr1_-_209792111 | 0.14 |
ENST00000455193.1
|
LAMB3
|
laminin, beta 3 |
| chr5_+_121647386 | 0.13 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr22_-_32766972 | 0.13 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr3_+_108308559 | 0.12 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr13_-_47471155 | 0.12 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr19_+_18303992 | 0.12 |
ENST00000599612.2
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr20_+_56964169 | 0.11 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr17_-_39150385 | 0.11 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr19_+_3762703 | 0.11 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr19_+_29456034 | 0.11 |
ENST00000589921.1
|
LINC00906
|
long intergenic non-protein coding RNA 906 |
| chr10_+_17270214 | 0.10 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr13_-_45048386 | 0.10 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chrX_+_73164149 | 0.10 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr2_-_31361543 | 0.10 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_+_44514040 | 0.10 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr12_-_10978957 | 0.10 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr12_+_54422142 | 0.10 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr10_+_135050908 | 0.09 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr20_+_5731083 | 0.09 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr12_-_52604607 | 0.09 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr14_+_68086515 | 0.09 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr7_-_55620433 | 0.09 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr9_-_128246769 | 0.09 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr20_+_56964253 | 0.09 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr10_-_29923893 | 0.09 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr7_+_107220899 | 0.09 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr6_+_147981838 | 0.09 |
ENST00000427015.1
ENST00000432506.1 |
RP11-307P5.1
|
RP11-307P5.1 |
| chr4_+_146539415 | 0.08 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr13_-_52733670 | 0.08 |
ENST00000550841.1
|
NEK3
|
NIMA-related kinase 3 |
| chr21_+_33671160 | 0.08 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr7_-_56101826 | 0.08 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr19_-_53758094 | 0.08 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr6_-_32095968 | 0.08 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr14_+_55493920 | 0.08 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr7_+_16793160 | 0.08 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr14_+_55494323 | 0.08 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr1_-_149908217 | 0.08 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr2_+_169312350 | 0.08 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr10_+_135204338 | 0.07 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr9_+_133539981 | 0.06 |
ENST00000253008.2
|
PRDM12
|
PR domain containing 12 |
| chr11_+_327171 | 0.06 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr3_-_180397256 | 0.06 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr2_+_158114051 | 0.06 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr1_+_46805832 | 0.06 |
ENST00000474844.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr8_+_30244580 | 0.06 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr6_-_32908765 | 0.06 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr14_+_55595762 | 0.06 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr6_-_32557610 | 0.06 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr15_+_67813406 | 0.06 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr4_-_164534657 | 0.05 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_-_114429997 | 0.05 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr14_-_64194745 | 0.05 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr10_+_13141585 | 0.05 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr5_+_67586465 | 0.05 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr5_-_75919217 | 0.05 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr7_+_7811992 | 0.05 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr12_-_11150474 | 0.05 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr6_-_32784687 | 0.05 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr15_-_42500351 | 0.05 |
ENST00000348544.4
ENST00000318006.5 |
VPS39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr14_+_52327109 | 0.05 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr14_+_77582905 | 0.04 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr2_+_175199674 | 0.04 |
ENST00000394967.2
|
SP9
|
Sp9 transcription factor |
| chr3_-_123512688 | 0.04 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123339418 | 0.04 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr6_+_28092338 | 0.04 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr10_-_101380121 | 0.04 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr13_+_50570019 | 0.04 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr6_-_109776901 | 0.04 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr1_-_160492994 | 0.04 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr19_-_7812446 | 0.04 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr10_+_13142225 | 0.04 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr10_-_43892668 | 0.04 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr12_+_122688090 | 0.04 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr8_+_28748765 | 0.04 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr7_-_20256965 | 0.04 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr11_-_85376121 | 0.04 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr3_-_47555167 | 0.03 |
ENST00000296149.4
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr15_-_77712477 | 0.03 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr1_-_19426149 | 0.03 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr15_+_51669513 | 0.03 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr11_+_92085262 | 0.03 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr14_+_55595960 | 0.03 |
ENST00000554715.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr2_-_158345462 | 0.03 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr20_+_5731027 | 0.03 |
ENST00000378979.4
ENST00000303142.6 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr18_+_29598335 | 0.03 |
ENST00000217740.3
|
RNF125
|
ring finger protein 125, E3 ubiquitin protein ligase |
| chr2_-_109605663 | 0.03 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr9_+_115913222 | 0.03 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chr22_+_41697520 | 0.03 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr19_-_7812397 | 0.03 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr2_-_88927092 | 0.02 |
ENST00000303236.3
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr6_+_131894284 | 0.02 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr16_-_66584059 | 0.02 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr18_-_47721447 | 0.02 |
ENST00000285039.7
|
MYO5B
|
myosin VB |
| chr11_+_22694123 | 0.02 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chr21_+_30502806 | 0.02 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr15_+_77712993 | 0.02 |
ENST00000336216.4
ENST00000381714.3 ENST00000558651.1 |
HMG20A
|
high mobility group 20A |
| chr1_+_84630645 | 0.02 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr20_+_34129770 | 0.02 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr17_-_48785216 | 0.02 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr1_-_109935819 | 0.02 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr7_+_141408153 | 0.02 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr8_-_99057742 | 0.02 |
ENST00000521291.1
ENST00000396070.2 ENST00000523172.1 ENST00000287038.3 |
RPL30
|
ribosomal protein L30 |
| chr14_-_75389925 | 0.02 |
ENST00000556776.1
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1 |
| chr5_+_82767487 | 0.02 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr5_+_102594403 | 0.02 |
ENST00000319933.2
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chr9_+_12695702 | 0.02 |
ENST00000381136.2
|
TYRP1
|
tyrosinase-related protein 1 |
| chr5_+_82767583 | 0.02 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr1_+_8021954 | 0.02 |
ENST00000377491.1
ENST00000377488.1 |
PARK7
|
parkinson protein 7 |
| chr4_-_186696425 | 0.02 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_55162453 | 0.02 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr5_-_137475071 | 0.01 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr15_-_77712429 | 0.01 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr17_-_8113542 | 0.01 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr1_-_72566613 | 0.01 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr16_-_66583701 | 0.01 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr9_-_23779367 | 0.01 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr5_+_175490540 | 0.01 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr18_-_70532906 | 0.01 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr9_-_86593238 | 0.01 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr14_+_50291993 | 0.01 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr5_+_121465207 | 0.01 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr11_+_45825616 | 0.01 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr14_-_73997901 | 0.01 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr4_+_119810134 | 0.01 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr7_-_36634181 | 0.01 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr1_+_160160283 | 0.01 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr5_+_82767284 | 0.01 |
ENST00000265077.3
|
VCAN
|
versican |
| chr6_-_109777128 | 0.01 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr5_+_140762268 | 0.00 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr21_+_39668478 | 0.00 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_-_147162078 | 0.00 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr15_-_99548775 | 0.00 |
ENST00000378919.6
|
PGPEP1L
|
pyroglutamyl-peptidase I-like |
| chr8_+_117963190 | 0.00 |
ENST00000427715.2
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr18_-_3874247 | 0.00 |
ENST00000581699.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr6_+_28249332 | 0.00 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr8_+_24241969 | 0.00 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr11_+_7506584 | 0.00 |
ENST00000530135.1
|
OLFML1
|
olfactomedin-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.2 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |