Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for MNT_HEY1_HEY2
Z-value: 0.60
Transcription factors associated with MNT_HEY1_HEY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
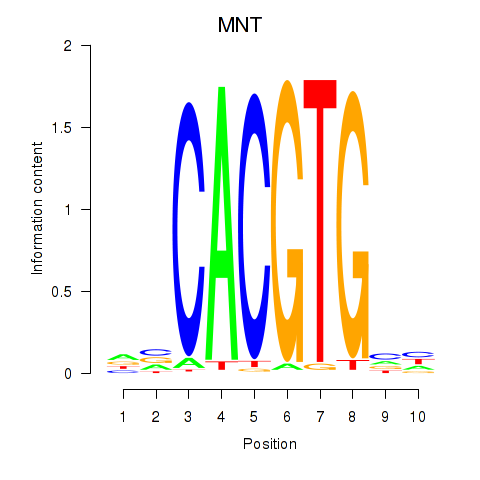
MNT
|
ENSG00000070444.10 | MAX network transcriptional repressor |
|
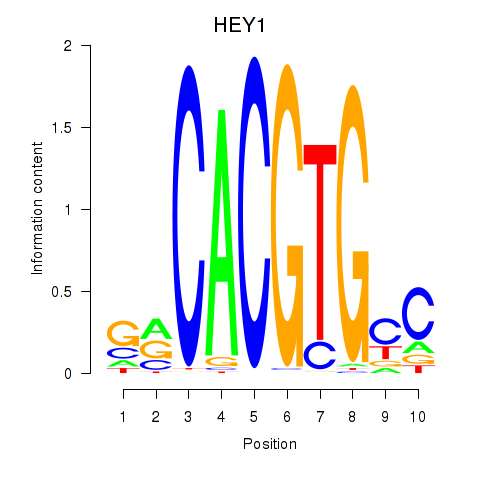
HEY1
|
ENSG00000164683.12 | hes related family bHLH transcription factor with YRPW motif 1 |
|
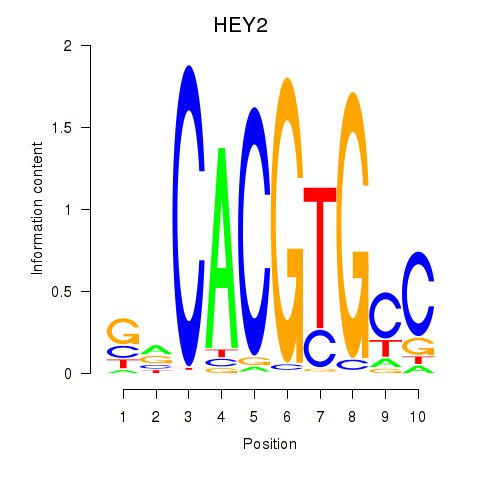
HEY2
|
ENSG00000135547.4 | hes related family bHLH transcription factor with YRPW motif 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HEY2 | hg19_v2_chr6_+_126070726_126070768 | -0.75 | 8.7e-02 | Click! |
| MNT | hg19_v2_chr17_-_2304365_2304412 | -0.43 | 4.0e-01 | Click! |
| HEY1 | hg19_v2_chr8_-_80680078_80680101 | -0.02 | 9.7e-01 | Click! |
Activity profile of MNT_HEY1_HEY2 motif
Sorted Z-values of MNT_HEY1_HEY2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_58145889 | 2.12 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr9_+_96928516 | 0.82 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr1_+_150254936 | 0.66 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr3_-_53080644 | 0.53 |
ENST00000497586.1
|
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr6_+_73331918 | 0.43 |
ENST00000402622.2
ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr3_+_52720187 | 0.37 |
ENST00000474423.1
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr5_-_133706695 | 0.35 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr16_+_2570431 | 0.31 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr21_-_46237883 | 0.30 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr2_-_232328867 | 0.28 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr16_+_66914264 | 0.27 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr3_-_196987309 | 0.26 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr10_+_104474353 | 0.26 |
ENST00000602647.1
ENST00000602439.1 ENST00000602764.1 |
SFXN2
|
sideroflexin 2 |
| chr10_-_126849626 | 0.26 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr17_-_35969409 | 0.25 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chr19_-_49016847 | 0.25 |
ENST00000598924.1
|
CTC-273B12.10
|
CTC-273B12.10 |
| chr1_-_26233423 | 0.24 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr2_-_198364581 | 0.24 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr3_-_53080672 | 0.24 |
ENST00000483069.1
|
SFMBT1
|
Scm-like with four mbt domains 1 |
| chrX_-_20134713 | 0.22 |
ENST00000452324.3
|
MAP7D2
|
MAP7 domain containing 2 |
| chr12_+_104235229 | 0.21 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein |
| chr2_+_198365095 | 0.21 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr10_-_44070016 | 0.21 |
ENST00000374446.2
ENST00000426961.1 ENST00000535642.1 |
ZNF239
|
zinc finger protein 239 |
| chr1_+_246729815 | 0.21 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chrX_-_20134990 | 0.20 |
ENST00000379651.3
ENST00000443379.3 ENST00000379643.5 |
MAP7D2
|
MAP7 domain containing 2 |
| chr2_+_240323439 | 0.20 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr1_-_26232951 | 0.20 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr15_+_89181974 | 0.20 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr5_-_154230130 | 0.19 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr2_-_27486951 | 0.19 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr4_+_17578815 | 0.19 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr15_+_27112948 | 0.19 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr6_-_30523865 | 0.19 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr3_-_178789220 | 0.19 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr6_-_33385823 | 0.18 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr14_-_105071083 | 0.18 |
ENST00000415614.2
|
TMEM179
|
transmembrane protein 179 |
| chr12_-_31478428 | 0.18 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr4_+_17579110 | 0.18 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr19_-_46974741 | 0.18 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr15_-_101817492 | 0.18 |
ENST00000528346.1
ENST00000531964.1 |
VIMP
|
VCP-interacting membrane protein |
| chr15_+_98503922 | 0.18 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr3_+_52719936 | 0.18 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr2_+_46926326 | 0.17 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr20_+_18118486 | 0.17 |
ENST00000432901.3
|
PET117
|
PET117 homolog (S. cerevisiae) |
| chr1_-_78149041 | 0.17 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr11_+_57435441 | 0.17 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr6_-_33385655 | 0.16 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_-_10694766 | 0.16 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr10_+_46222648 | 0.16 |
ENST00000336378.4
ENST00000540872.1 ENST00000537517.1 ENST00000374362.2 ENST00000359860.4 ENST00000420848.1 |
FAM21C
|
family with sequence similarity 21, member C |
| chr3_-_52719810 | 0.16 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr3_-_69062742 | 0.16 |
ENST00000424374.1
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_-_126076264 | 0.15 |
ENST00000296233.3
|
KLF15
|
Kruppel-like factor 15 |
| chr3_+_122785895 | 0.15 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr8_+_75896849 | 0.15 |
ENST00000520277.1
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chrX_-_154255143 | 0.15 |
ENST00000453950.1
ENST00000423959.1 |
F8
|
coagulation factor VIII, procoagulant component |
| chr17_+_48503519 | 0.15 |
ENST00000300441.4
ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr17_-_42907564 | 0.15 |
ENST00000592524.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr6_-_13486369 | 0.15 |
ENST00000558378.1
|
AL583828.1
|
AL583828.1 |
| chr17_+_40985407 | 0.15 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr10_+_180405 | 0.15 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr11_+_7534999 | 0.15 |
ENST00000528947.1
ENST00000299492.4 |
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_64845864 | 0.15 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr3_+_186649133 | 0.14 |
ENST00000417392.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr12_+_104359614 | 0.14 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr1_-_241520525 | 0.14 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr6_+_138188351 | 0.14 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr2_+_46926048 | 0.14 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr8_-_54755789 | 0.14 |
ENST00000359530.2
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr9_+_37079968 | 0.14 |
ENST00000588403.1
|
RP11-220I1.1
|
RP11-220I1.1 |
| chr18_-_21977748 | 0.13 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr3_+_133292574 | 0.13 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr2_-_232329186 | 0.13 |
ENST00000322723.4
|
NCL
|
nucleolin |
| chrX_-_57163430 | 0.13 |
ENST00000374908.1
|
SPIN2A
|
spindlin family, member 2A |
| chr9_-_100684769 | 0.13 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chrX_-_34675391 | 0.13 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr12_+_9102632 | 0.13 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr8_+_17354617 | 0.13 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr12_-_90103077 | 0.13 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr22_-_36903069 | 0.12 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr17_-_6915616 | 0.12 |
ENST00000575889.1
|
AC027763.2
|
Uncharacterized protein |
| chr14_+_103394963 | 0.12 |
ENST00000559525.1
ENST00000559789.1 |
AMN
|
amnion associated transmembrane protein |
| chr12_+_64845660 | 0.12 |
ENST00000331710.5
|
TBK1
|
TANK-binding kinase 1 |
| chr5_-_122759032 | 0.12 |
ENST00000510582.3
ENST00000328236.5 ENST00000306481.6 ENST00000508442.2 ENST00000395431.2 |
CEP120
|
centrosomal protein 120kDa |
| chr17_+_27052892 | 0.12 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr11_+_120894781 | 0.12 |
ENST00000529397.1
ENST00000528512.1 ENST00000422003.2 |
TBCEL
|
tubulin folding cofactor E-like |
| chr17_-_34890709 | 0.12 |
ENST00000544606.1
|
MYO19
|
myosin XIX |
| chr6_+_151561506 | 0.12 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_+_198365122 | 0.12 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr13_+_27998681 | 0.12 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr7_+_23636992 | 0.12 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr1_-_154193009 | 0.12 |
ENST00000368518.1
ENST00000368519.1 ENST00000368521.5 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr5_-_158636512 | 0.12 |
ENST00000424310.2
|
RNF145
|
ring finger protein 145 |
| chr7_-_44365216 | 0.12 |
ENST00000358707.3
ENST00000457475.1 ENST00000440254.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr4_+_57302297 | 0.11 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr15_+_96875657 | 0.11 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_148778570 | 0.11 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr11_+_45944190 | 0.11 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr14_-_75735986 | 0.11 |
ENST00000553510.1
|
RP11-293M10.1
|
Uncharacterized protein |
| chr3_+_133292851 | 0.11 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr16_-_4588391 | 0.11 |
ENST00000586728.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr1_-_151319318 | 0.11 |
ENST00000436271.1
ENST00000450506.1 ENST00000422595.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr17_-_46688334 | 0.11 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr17_+_48503603 | 0.11 |
ENST00000502667.1
|
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr14_-_20923195 | 0.11 |
ENST00000206542.4
|
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr9_+_6758109 | 0.11 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr11_-_119252425 | 0.11 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr11_-_119252359 | 0.11 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr1_+_28261533 | 0.11 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr7_-_102985035 | 0.11 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr4_+_183065793 | 0.11 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr9_+_129987488 | 0.11 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr1_+_154193325 | 0.11 |
ENST00000428931.1
ENST00000441890.1 ENST00000271877.7 ENST00000412596.1 ENST00000368504.1 ENST00000437652.1 |
UBAP2L
|
ubiquitin associated protein 2-like |
| chr9_+_74526532 | 0.11 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr17_+_74723031 | 0.11 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr3_-_52719546 | 0.10 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr19_-_49137790 | 0.10 |
ENST00000599385.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr17_-_36831156 | 0.10 |
ENST00000325814.5
|
C17orf96
|
chromosome 17 open reading frame 96 |
| chr12_-_122750957 | 0.10 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr13_-_52027134 | 0.10 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr1_-_26232522 | 0.10 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr19_-_14316980 | 0.10 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr7_+_23637118 | 0.10 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr6_+_151561085 | 0.10 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr6_+_151646800 | 0.10 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr1_+_43637996 | 0.10 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr7_-_27135591 | 0.10 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr1_-_6259613 | 0.10 |
ENST00000465387.1
|
RPL22
|
ribosomal protein L22 |
| chr18_-_33077942 | 0.10 |
ENST00000334598.7
|
INO80C
|
INO80 complex subunit C |
| chr17_-_34890665 | 0.10 |
ENST00000586007.1
|
MYO19
|
myosin XIX |
| chr7_-_44365020 | 0.10 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr17_-_17942473 | 0.10 |
ENST00000585101.1
ENST00000474627.3 ENST00000444058.1 |
ATPAF2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr2_-_68384603 | 0.10 |
ENST00000406245.2
ENST00000409164.1 ENST00000295121.6 |
WDR92
|
WD repeat domain 92 |
| chr12_-_15374343 | 0.10 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr7_-_32931623 | 0.10 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr12_+_50355647 | 0.10 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr12_+_104359641 | 0.10 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr17_+_49337881 | 0.10 |
ENST00000225298.7
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast) |
| chr1_-_159915386 | 0.10 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr16_+_3704822 | 0.10 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr10_-_16859361 | 0.10 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr3_+_133292759 | 0.10 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr15_+_89182156 | 0.10 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr17_+_76311791 | 0.10 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr8_+_109455845 | 0.10 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr22_+_45680822 | 0.10 |
ENST00000216211.4
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr17_-_5372068 | 0.10 |
ENST00000572490.1
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr2_+_27805971 | 0.10 |
ENST00000413371.2
|
ZNF512
|
zinc finger protein 512 |
| chr4_+_76439095 | 0.10 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr2_-_119605253 | 0.10 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr3_-_189839467 | 0.10 |
ENST00000426003.1
|
LEPREL1
|
leprecan-like 1 |
| chr6_+_144164455 | 0.09 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr1_-_231376867 | 0.09 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr15_+_89182178 | 0.09 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr7_-_69062391 | 0.09 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chrX_+_10126488 | 0.09 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr8_+_133787586 | 0.09 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr1_+_150293921 | 0.09 |
ENST00000324862.6
|
PRPF3
|
pre-mRNA processing factor 3 |
| chr3_+_184530173 | 0.09 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr8_-_144442094 | 0.09 |
ENST00000521193.1
ENST00000520950.1 |
TOP1MT
|
topoisomerase (DNA) I, mitochondrial |
| chr17_-_7197881 | 0.09 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chr6_+_138188378 | 0.09 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr1_+_45140400 | 0.09 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr2_+_86333340 | 0.09 |
ENST00000409783.2
ENST00000409277.3 |
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr17_-_73975198 | 0.09 |
ENST00000301608.4
ENST00000588176.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr1_-_45956868 | 0.09 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr8_+_17354587 | 0.09 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr14_+_52780998 | 0.09 |
ENST00000557436.1
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa |
| chr2_+_187350973 | 0.09 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr1_-_42921915 | 0.09 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr2_+_201171372 | 0.09 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chrX_+_108780347 | 0.09 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr9_+_37079888 | 0.09 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr12_-_6715808 | 0.09 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr7_-_75677251 | 0.09 |
ENST00000431581.1
ENST00000359697.3 ENST00000451157.1 ENST00000340062.5 ENST00000360591.3 ENST00000248600.1 |
STYXL1
|
serine/threonine/tyrosine interacting-like 1 |
| chr13_+_103451399 | 0.09 |
ENST00000257336.1
ENST00000448849.2 |
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr11_-_64764435 | 0.09 |
ENST00000534177.1
ENST00000301887.4 |
BATF2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr17_-_5342380 | 0.09 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr4_-_146019335 | 0.09 |
ENST00000451299.2
ENST00000507656.1 ENST00000309439.5 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr1_-_231376836 | 0.09 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr20_-_2821271 | 0.09 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr17_-_47755338 | 0.09 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr17_-_5372229 | 0.09 |
ENST00000433302.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr18_-_33077556 | 0.08 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr17_-_49021974 | 0.08 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr17_+_73975292 | 0.08 |
ENST00000397640.1
ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 CST complex subunit |
| chr5_+_80256453 | 0.08 |
ENST00000265080.4
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2 |
| chr1_+_43148625 | 0.08 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr16_-_4588469 | 0.08 |
ENST00000588381.1
ENST00000563332.2 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr2_+_44396000 | 0.08 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr9_-_131709858 | 0.08 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr17_+_5185552 | 0.08 |
ENST00000262477.6
ENST00000408982.2 ENST00000575991.1 ENST00000537505.1 ENST00000546142.2 |
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr8_-_54755459 | 0.08 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr2_-_11810284 | 0.08 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr2_+_176957619 | 0.08 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chrX_-_13956737 | 0.08 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chr21_-_16374688 | 0.08 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr3_+_127317705 | 0.08 |
ENST00000480910.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr16_-_28634874 | 0.08 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr9_+_130890612 | 0.08 |
ENST00000443493.1
|
AL590708.2
|
AL590708.2 |
| chr22_+_46972975 | 0.08 |
ENST00000431155.1
|
GRAMD4
|
GRAM domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MNT_HEY1_HEY2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.3 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.6 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.2 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 | 0.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.1 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.0 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.0 | 0.0 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.6 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.2 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.0 | GO:0006664 | glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0009757 | hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.0 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.0 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of mitotic centrosome separation(GO:0046604) positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.0 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.0 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.0 | 0.0 | GO:0061184 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) |
| 0.0 | 0.0 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.0 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.0 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.0 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.0 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.0 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.0 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.2 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule membrane(GO:0031088) platelet dense granule(GO:0042827) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.0 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.0 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.2 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.1 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 2.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.0 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.0 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |