Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
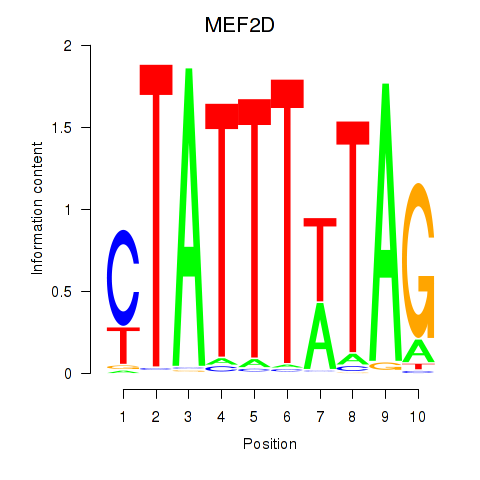
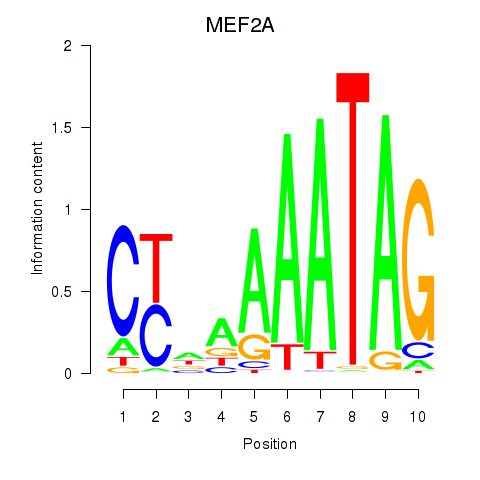
Results for MEF2D_MEF2A
Z-value: 0.28
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2D
|
ENSG00000116604.13 | myocyte enhancer factor 2D |
|
MEF2A
|
ENSG00000068305.13 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2A | hg19_v2_chr15_+_100106155_100106243 | 0.91 | 1.1e-02 | Click! |
| MEF2D | hg19_v2_chr1_-_156470515_156470542 | -0.80 | 5.6e-02 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_111091948 | 0.37 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr14_+_56046990 | 0.25 |
ENST00000438792.2
ENST00000395314.3 ENST00000395308.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr6_-_6007200 | 0.24 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr11_+_12108410 | 0.24 |
ENST00000527997.1
|
RP13-631K18.5
|
RP13-631K18.5 |
| chr14_+_58797974 | 0.24 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_-_148779106 | 0.24 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr7_-_124569991 | 0.20 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr7_-_124569864 | 0.20 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr9_-_104145795 | 0.19 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr14_+_56046914 | 0.19 |
ENST00000413890.2
ENST00000395309.3 ENST00000554567.1 ENST00000555498.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr15_+_100106155 | 0.17 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr2_-_27938593 | 0.17 |
ENST00000379677.2
|
AC074091.13
|
Uncharacterized protein |
| chr3_-_148939598 | 0.17 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_+_90098606 | 0.17 |
ENST00000370454.4
|
LRRC8C
|
leucine rich repeat containing 8 family, member C |
| chr6_+_151358048 | 0.16 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr8_-_95565673 | 0.16 |
ENST00000437199.1
ENST00000297591.5 ENST00000421249.2 |
KIAA1429
|
KIAA1429 |
| chr4_-_143767428 | 0.15 |
ENST00000513000.1
ENST00000509777.1 ENST00000503927.1 |
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr9_-_119162885 | 0.15 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr1_-_117753540 | 0.15 |
ENST00000328189.3
ENST00000369458.3 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr8_+_107282389 | 0.14 |
ENST00000577661.1
ENST00000445937.1 |
RP11-395G23.3
OXR1
|
RP11-395G23.3 oxidation resistance 1 |
| chr15_-_70994612 | 0.14 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr14_+_39735411 | 0.14 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr11_+_65266507 | 0.14 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr2_+_103378472 | 0.14 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr2_-_148778323 | 0.13 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr7_-_35013217 | 0.13 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr1_-_204436344 | 0.13 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr15_+_100106244 | 0.13 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr12_+_40618764 | 0.12 |
ENST00000343742.2
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr4_+_95373037 | 0.12 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr6_+_123038689 | 0.12 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr6_+_132129151 | 0.12 |
ENST00000360971.2
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr3_-_145878954 | 0.12 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr21_+_35107346 | 0.11 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr9_-_33402506 | 0.11 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr4_+_57276661 | 0.11 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr1_-_150693318 | 0.11 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr1_-_100643765 | 0.11 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chrM_+_7586 | 0.11 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_-_151217166 | 0.11 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr11_-_7904464 | 0.11 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr5_+_138609782 | 0.11 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr1_+_150254936 | 0.11 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr2_-_208030886 | 0.11 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr2_+_44502597 | 0.10 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chrX_-_21776281 | 0.10 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr8_+_27631903 | 0.10 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr2_+_145780725 | 0.10 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr2_-_148778258 | 0.10 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr1_+_25757376 | 0.10 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr2_+_161993465 | 0.10 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr8_-_17533838 | 0.09 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr3_+_138067521 | 0.09 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr8_-_70745575 | 0.09 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chrX_-_128657457 | 0.09 |
ENST00000371121.3
ENST00000371123.1 ENST00000371122.4 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr2_-_214017151 | 0.09 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr10_-_14880002 | 0.09 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chrY_-_23548246 | 0.09 |
ENST00000382764.1
|
CYorf17
|
chromosome Y open reading frame 17 |
| chr20_+_5987890 | 0.09 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr3_-_148939835 | 0.08 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr3_-_176914238 | 0.08 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr18_-_64271316 | 0.08 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr5_-_137475071 | 0.08 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr3_-_192445289 | 0.08 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr17_-_36891830 | 0.08 |
ENST00000578487.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr4_+_55095428 | 0.08 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chrX_+_119384607 | 0.08 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
| chr3_+_50316458 | 0.08 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr3_-_108248169 | 0.08 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr16_+_31225337 | 0.08 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr2_+_161993412 | 0.08 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr17_+_37821593 | 0.08 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr22_-_41258074 | 0.08 |
ENST00000307221.4
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr6_-_76203345 | 0.08 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr2_+_220299547 | 0.07 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr17_+_7184986 | 0.07 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr11_-_65837090 | 0.07 |
ENST00000529036.1
|
RP11-1167A19.2
|
Uncharacterized protein |
| chr11_-_47400062 | 0.07 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr12_+_20963647 | 0.07 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr2_+_33683109 | 0.07 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr12_+_69201923 | 0.07 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr1_-_150693305 | 0.07 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr3_+_99833755 | 0.07 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr7_-_29152509 | 0.07 |
ENST00000448959.1
|
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr7_+_30951461 | 0.07 |
ENST00000311813.4
|
AQP1
|
aquaporin 1 (Colton blood group) |
| chr12_+_110562135 | 0.07 |
ENST00000361948.4
ENST00000552912.1 ENST00000242591.5 ENST00000546374.1 |
IFT81
|
intraflagellar transport 81 homolog (Chlamydomonas) |
| chr14_+_56078695 | 0.07 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr2_+_145780739 | 0.07 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr6_+_136172820 | 0.07 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr15_-_52944231 | 0.07 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr11_-_47399942 | 0.07 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr17_-_38821373 | 0.07 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_-_63782888 | 0.07 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr6_-_76203454 | 0.07 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr17_+_68165657 | 0.07 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr15_+_74509530 | 0.06 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chrX_+_123097014 | 0.06 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr9_+_8858102 | 0.06 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr2_-_179914760 | 0.06 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr18_-_64271363 | 0.06 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr3_-_27498235 | 0.06 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr22_-_36018569 | 0.06 |
ENST00000419229.1
ENST00000406324.1 |
MB
|
myoglobin |
| chr8_+_40010989 | 0.06 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr3_+_121311966 | 0.06 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr10_+_14880157 | 0.06 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr8_+_76452097 | 0.06 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr13_+_76378305 | 0.06 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr18_+_9708162 | 0.06 |
ENST00000578921.1
|
RAB31
|
RAB31, member RAS oncogene family |
| chr8_+_74903580 | 0.06 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr4_+_119809984 | 0.06 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr19_-_51220176 | 0.06 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr12_+_20963632 | 0.06 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr4_-_39640513 | 0.06 |
ENST00000511809.1
ENST00000505729.1 |
SMIM14
|
small integral membrane protein 14 |
| chr2_-_165424973 | 0.06 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr6_+_159291090 | 0.06 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr1_+_92632542 | 0.06 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr5_-_66492562 | 0.05 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr7_+_7196565 | 0.05 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr7_+_47694842 | 0.05 |
ENST00000408988.2
|
C7orf65
|
chromosome 7 open reading frame 65 |
| chrX_+_591524 | 0.05 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr11_-_47400078 | 0.05 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr18_+_3252265 | 0.05 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_-_200379180 | 0.05 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr11_+_1860200 | 0.05 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr3_+_186739636 | 0.05 |
ENST00000440338.1
ENST00000448044.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr17_-_41132088 | 0.05 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr19_-_47975106 | 0.05 |
ENST00000539381.1
ENST00000594353.1 ENST00000542837.1 |
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr17_-_39211463 | 0.05 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr4_+_129730779 | 0.05 |
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1 |
| chr2_+_145780767 | 0.05 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr1_-_27240455 | 0.05 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr21_-_15583165 | 0.05 |
ENST00000536861.1
|
LIPI
|
lipase, member I |
| chr6_-_134373732 | 0.05 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr14_-_62217779 | 0.05 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr16_-_18923035 | 0.05 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr22_+_31742875 | 0.05 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr6_+_159290917 | 0.05 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr7_+_120629653 | 0.05 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr17_-_6917755 | 0.05 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr1_-_917466 | 0.05 |
ENST00000341290.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr2_-_151344172 | 0.05 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr5_+_155753745 | 0.04 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr20_+_9049682 | 0.04 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr7_+_18126557 | 0.04 |
ENST00000417496.2
|
HDAC9
|
histone deacetylase 9 |
| chr6_-_135424186 | 0.04 |
ENST00000529882.1
|
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr9_-_15472730 | 0.04 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr15_+_78558523 | 0.04 |
ENST00000446172.2
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr1_+_84630574 | 0.04 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_-_45269430 | 0.04 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr4_+_89514402 | 0.04 |
ENST00000426683.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr6_+_131571535 | 0.04 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr13_-_50367057 | 0.04 |
ENST00000261667.3
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4) |
| chr19_+_45973120 | 0.04 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr12_-_16760021 | 0.04 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr5_+_66124590 | 0.04 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_203144941 | 0.04 |
ENST00000255416.4
|
MYBPH
|
myosin binding protein H |
| chr11_+_57308979 | 0.04 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr6_+_26124373 | 0.04 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chrX_-_77914825 | 0.04 |
ENST00000321110.1
|
ZCCHC5
|
zinc finger, CCHC domain containing 5 |
| chr10_+_86184676 | 0.04 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr13_+_76378357 | 0.04 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr10_+_32856764 | 0.04 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr3_+_52448539 | 0.04 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr13_+_51483814 | 0.04 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr12_+_116985896 | 0.04 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr2_-_218766698 | 0.04 |
ENST00000439083.1
|
TNS1
|
tensin 1 |
| chr17_+_65040678 | 0.04 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr17_+_80186908 | 0.04 |
ENST00000582743.1
ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr1_-_31845914 | 0.04 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr2_+_44502630 | 0.04 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr4_-_25865159 | 0.04 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr9_+_100263912 | 0.04 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr15_+_40531621 | 0.04 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_+_65663812 | 0.04 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr7_+_143013198 | 0.04 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr8_-_66701319 | 0.04 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr1_+_95583479 | 0.03 |
ENST00000455656.1
ENST00000604534.1 |
TMEM56
RP11-57H12.6
|
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr3_+_69928256 | 0.03 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr15_-_99057551 | 0.03 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chrX_+_123095860 | 0.03 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr17_+_7461613 | 0.03 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr8_-_74659693 | 0.03 |
ENST00000518767.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr8_-_42358742 | 0.03 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr18_+_3252206 | 0.03 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_+_170366203 | 0.03 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr12_+_14561422 | 0.03 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr1_-_28384598 | 0.03 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chrX_-_133792480 | 0.03 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr16_+_84328252 | 0.03 |
ENST00000219454.5
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr19_-_47128294 | 0.03 |
ENST00000596260.1
ENST00000597185.1 ENST00000598865.1 ENST00000594275.1 ENST00000291294.2 |
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP) |
| chr19_+_35630344 | 0.03 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr4_+_55096010 | 0.03 |
ENST00000503856.1
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr17_-_74380565 | 0.03 |
ENST00000442767.1
ENST00000423915.1 |
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
| chr3_+_46924829 | 0.03 |
ENST00000313049.5
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr1_+_26485511 | 0.03 |
ENST00000374268.3
|
FAM110D
|
family with sequence similarity 110, member D |
| chr18_+_42260059 | 0.03 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr12_+_44229846 | 0.03 |
ENST00000551577.1
ENST00000266534.3 |
TMEM117
|
transmembrane protein 117 |
| chr3_+_135741576 | 0.03 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chrX_-_63450480 | 0.03 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr1_+_84630645 | 0.03 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.2 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.1 | GO:1902823 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0044027 | DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |