Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
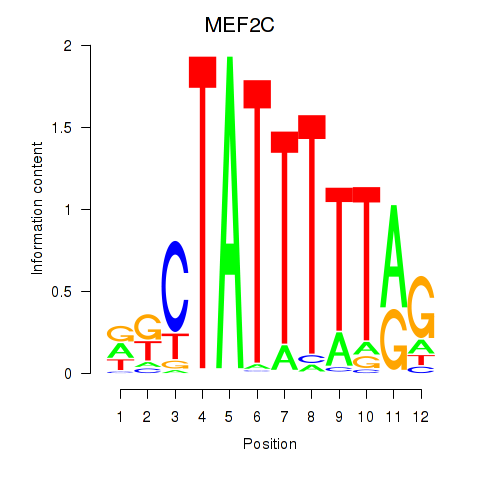
Results for MEF2C
Z-value: 0.85
Transcription factors associated with MEF2C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2C
|
ENSG00000081189.9 | myocyte enhancer factor 2C |
Activity profile of MEF2C motif
Sorted Z-values of MEF2C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_65837090 | 0.87 |
ENST00000529036.1
|
RP11-1167A19.2
|
Uncharacterized protein |
| chr8_+_101349823 | 0.76 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr17_+_7758374 | 0.59 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chrX_+_153046456 | 0.47 |
ENST00000393786.3
ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3
|
SRSF protein kinase 3 |
| chr3_+_193853927 | 0.45 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr3_+_8543393 | 0.44 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr9_-_102582155 | 0.40 |
ENST00000427039.1
|
RP11-554F20.1
|
RP11-554F20.1 |
| chr21_+_17909594 | 0.40 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_206945830 | 0.40 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chrX_+_9217932 | 0.38 |
ENST00000432442.1
|
GS1-519E5.1
|
GS1-519E5.1 |
| chr7_-_44105158 | 0.37 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr11_-_62457371 | 0.37 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr3_-_4927447 | 0.33 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr17_-_27467418 | 0.32 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr17_-_72855989 | 0.32 |
ENST00000293190.5
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
| chr4_+_37892682 | 0.31 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr19_-_49926698 | 0.29 |
ENST00000270631.1
|
PTH2
|
parathyroid hormone 2 |
| chr4_-_185303418 | 0.29 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr2_+_239047337 | 0.29 |
ENST00000409223.1
ENST00000305959.4 |
KLHL30
|
kelch-like family member 30 |
| chrX_-_15683147 | 0.29 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr10_-_45474237 | 0.28 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr2_+_30569506 | 0.27 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr19_-_49576198 | 0.27 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr3_-_137834436 | 0.27 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr3_+_8543533 | 0.26 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr12_+_50690489 | 0.25 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr8_-_70745575 | 0.25 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr16_-_85969774 | 0.25 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr15_+_59730348 | 0.25 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr17_+_37821593 | 0.24 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr12_-_371994 | 0.24 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr17_-_42200996 | 0.24 |
ENST00000587135.1
ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5
|
histone deacetylase 5 |
| chr3_-_52486841 | 0.24 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr16_+_12059091 | 0.23 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr22_+_45680822 | 0.23 |
ENST00000216211.4
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr2_+_145780662 | 0.23 |
ENST00000423031.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr17_-_79881408 | 0.23 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr4_+_89514516 | 0.23 |
ENST00000452979.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr12_+_116985896 | 0.22 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr16_+_14280564 | 0.22 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr11_+_19799327 | 0.22 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr3_+_8543561 | 0.21 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chrX_+_123095860 | 0.21 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr8_+_95565947 | 0.21 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr11_-_47374246 | 0.20 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr22_-_48943199 | 0.20 |
ENST00000407505.3
|
CTA-299D3.8
|
Uncharacterized protein |
| chr7_-_105332084 | 0.20 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr17_-_42200958 | 0.19 |
ENST00000336057.5
|
HDAC5
|
histone deacetylase 5 |
| chr8_-_73793975 | 0.19 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr15_+_42651691 | 0.19 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr2_-_218766698 | 0.19 |
ENST00000439083.1
|
TNS1
|
tensin 1 |
| chr8_+_86999516 | 0.19 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr1_+_156095951 | 0.18 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr1_+_228395755 | 0.18 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr20_+_32581525 | 0.18 |
ENST00000246194.3
ENST00000413297.1 |
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr20_+_32581738 | 0.17 |
ENST00000333552.5
|
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr1_-_156460391 | 0.17 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chrX_-_15332665 | 0.17 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr6_+_123038689 | 0.17 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr17_+_7184986 | 0.17 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr12_-_122240792 | 0.17 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr1_-_12677714 | 0.17 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr22_-_36031181 | 0.16 |
ENST00000594060.1
|
AL049747.1
|
AL049747.1 |
| chr13_-_45048386 | 0.16 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr13_-_30880979 | 0.16 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr8_+_76452097 | 0.16 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr9_+_70971815 | 0.15 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr2_-_100925967 | 0.15 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr3_+_50316458 | 0.14 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr16_+_14280742 | 0.14 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr18_-_19283649 | 0.14 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr4_+_113970772 | 0.14 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr2_+_109204909 | 0.14 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_-_143226979 | 0.14 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chrX_-_30327495 | 0.14 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr11_-_47400032 | 0.14 |
ENST00000533968.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr7_+_18535346 | 0.13 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr15_-_52970820 | 0.13 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr8_-_27115903 | 0.13 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr2_-_86564696 | 0.13 |
ENST00000437769.1
|
REEP1
|
receptor accessory protein 1 |
| chr4_-_23891693 | 0.13 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr2_-_179914760 | 0.13 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr9_-_110251836 | 0.13 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr6_+_15401075 | 0.13 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr2_+_220144052 | 0.13 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr4_+_89514402 | 0.13 |
ENST00000426683.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr16_+_50313426 | 0.13 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr3_+_179370517 | 0.13 |
ENST00000263966.3
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr7_+_18535321 | 0.13 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr3_-_30936153 | 0.13 |
ENST00000454381.3
ENST00000282538.5 |
GADL1
|
glutamate decarboxylase-like 1 |
| chr19_+_35630344 | 0.13 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_+_221051699 | 0.12 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr1_+_104104379 | 0.12 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr2_+_220144168 | 0.12 |
ENST00000392087.2
ENST00000442681.1 ENST00000439026.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_+_156096336 | 0.12 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr4_-_15661474 | 0.12 |
ENST00000509314.1
ENST00000503196.1 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr8_-_33457453 | 0.12 |
ENST00000523956.1
ENST00000256261.4 |
DUSP26
|
dual specificity phosphatase 26 (putative) |
| chr5_+_173316341 | 0.12 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_+_201979743 | 0.12 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr12_-_70004942 | 0.12 |
ENST00000361484.3
|
LRRC10
|
leucine rich repeat containing 10 |
| chr2_+_220143989 | 0.12 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr7_+_18535893 | 0.11 |
ENST00000432645.2
ENST00000441542.2 |
HDAC9
|
histone deacetylase 9 |
| chr2_-_237172988 | 0.11 |
ENST00000409749.3
ENST00000430053.1 |
ASB18
|
ankyrin repeat and SOCS box containing 18 |
| chr7_-_124569864 | 0.11 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr9_+_127054217 | 0.11 |
ENST00000394199.2
ENST00000546191.1 |
NEK6
|
NIMA-related kinase 6 |
| chr1_-_201390846 | 0.11 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chrX_+_18725758 | 0.11 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr1_-_230850043 | 0.11 |
ENST00000366667.4
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr3_+_4535025 | 0.11 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr9_+_116327326 | 0.11 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr5_+_140749803 | 0.10 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chrX_-_138287168 | 0.10 |
ENST00000436198.1
|
FGF13
|
fibroblast growth factor 13 |
| chr3_+_159570722 | 0.10 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr9_+_5231413 | 0.10 |
ENST00000239316.4
|
INSL4
|
insulin-like 4 (placenta) |
| chr15_+_78558523 | 0.10 |
ENST00000446172.2
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr10_-_75410771 | 0.10 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr10_-_29923893 | 0.09 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr1_+_201979645 | 0.09 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr8_-_94029882 | 0.09 |
ENST00000520686.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr4_-_143227088 | 0.09 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr10_-_75415825 | 0.09 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr15_+_67418047 | 0.09 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr2_+_65663812 | 0.09 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr2_+_179184955 | 0.09 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr7_-_82073109 | 0.09 |
ENST00000356860.3
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr7_-_55583740 | 0.09 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr1_+_198608146 | 0.08 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr1_-_154164534 | 0.08 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr14_+_39735411 | 0.08 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr8_+_107282368 | 0.08 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr3_+_135741576 | 0.08 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr4_-_23891658 | 0.08 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr1_-_204436344 | 0.08 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr12_-_118406028 | 0.08 |
ENST00000425217.1
|
KSR2
|
kinase suppressor of ras 2 |
| chr18_-_35145981 | 0.08 |
ENST00000420428.2
ENST00000412753.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr19_-_39340563 | 0.08 |
ENST00000601813.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr3_+_138067666 | 0.08 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr7_-_22862406 | 0.08 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr13_-_67802549 | 0.08 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr2_-_151344172 | 0.08 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr7_+_152456904 | 0.08 |
ENST00000537264.1
|
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr13_+_111766897 | 0.08 |
ENST00000317133.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr5_-_142065612 | 0.08 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr13_-_30881134 | 0.08 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr4_+_39408470 | 0.08 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr1_+_203097407 | 0.08 |
ENST00000367235.1
|
ADORA1
|
adenosine A1 receptor |
| chr1_+_183605222 | 0.08 |
ENST00000536277.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr1_+_201617264 | 0.07 |
ENST00000367296.4
|
NAV1
|
neuron navigator 1 |
| chr14_+_39736582 | 0.07 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr15_-_76352069 | 0.07 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr17_-_45899126 | 0.07 |
ENST00000007414.3
ENST00000392507.3 |
OSBPL7
|
oxysterol binding protein-like 7 |
| chr3_+_113616317 | 0.07 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr2_+_109204743 | 0.07 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_-_34524157 | 0.07 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr9_-_215744 | 0.07 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr8_+_132952112 | 0.07 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr18_+_29027696 | 0.07 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr17_+_7461580 | 0.07 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr22_+_41487711 | 0.07 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr17_+_17685422 | 0.07 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr2_-_220117867 | 0.07 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr1_+_201617450 | 0.07 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr4_+_129730839 | 0.07 |
ENST00000511647.1
|
PHF17
|
jade family PHD finger 1 |
| chr5_-_131347306 | 0.06 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr10_-_104179682 | 0.06 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr11_+_65266507 | 0.06 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr20_+_3767547 | 0.06 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr10_-_104178857 | 0.06 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr3_-_176914191 | 0.06 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr9_+_127054254 | 0.06 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr9_+_215158 | 0.06 |
ENST00000479404.1
|
DOCK8
|
dedicator of cytokinesis 8 |
| chr1_-_40042416 | 0.06 |
ENST00000372857.3
ENST00000372856.3 ENST00000531243.2 ENST00000451091.2 |
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr1_-_204329013 | 0.06 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr15_+_100106244 | 0.06 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr11_+_128563652 | 0.06 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_+_37545012 | 0.06 |
ENST00000378616.3
|
XK
|
X-linked Kx blood group (McLeod syndrome) |
| chr17_+_65040678 | 0.06 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr14_+_74003818 | 0.06 |
ENST00000311148.4
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr11_+_7598239 | 0.06 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_53846612 | 0.06 |
ENST00000551104.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr7_+_129906660 | 0.06 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr1_-_24438664 | 0.06 |
ENST00000374434.3
ENST00000330966.7 ENST00000329601.7 |
MYOM3
|
myomesin 3 |
| chr4_+_120056939 | 0.06 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr1_-_226187013 | 0.05 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr10_-_61900762 | 0.05 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr18_-_3219847 | 0.05 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr3_+_127634069 | 0.05 |
ENST00000405109.1
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr1_+_24018269 | 0.05 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr9_+_130565147 | 0.05 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr8_+_145162629 | 0.05 |
ENST00000323662.8
|
KIAA1875
|
KIAA1875 |
| chr20_+_58251716 | 0.05 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_-_22862448 | 0.05 |
ENST00000358435.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr15_-_93353028 | 0.05 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr3_+_127634312 | 0.05 |
ENST00000407609.3
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr10_-_62704005 | 0.05 |
ENST00000337910.5
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr11_-_116708302 | 0.05 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr4_-_71532601 | 0.05 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr10_+_102758105 | 0.05 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr17_-_27949911 | 0.05 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr8_-_139926236 | 0.05 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.4 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.2 | GO:1904640 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.1 | 0.5 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.3 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0061461 | angiotensin-mediated drinking behavior(GO:0003051) lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.1 | GO:1904995 | regulation of muscle hyperplasia(GO:0014738) negative regulation of muscle hyperplasia(GO:0014740) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0021564 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.4 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:2000544 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |