Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
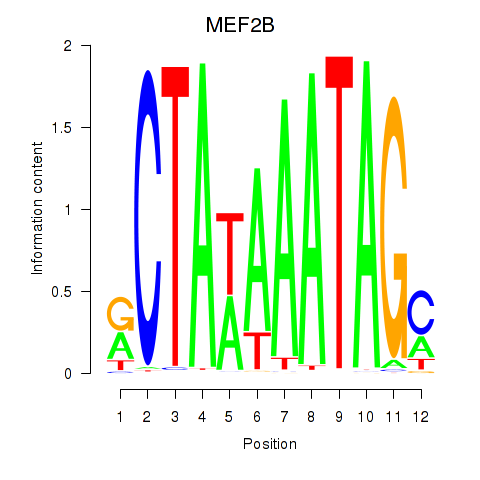
Results for MEF2B
Z-value: 0.43
Transcription factors associated with MEF2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2B
|
ENSG00000213999.11 | myocyte enhancer factor 2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2B | hg19_v2_chr19_-_19281054_19281098 | 0.88 | 2.2e-02 | Click! |
Activity profile of MEF2B motif
Sorted Z-values of MEF2B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_107282368 | 0.28 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr3_-_188665428 | 0.24 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr22_-_30642728 | 0.21 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr12_+_54384370 | 0.19 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr22_+_19706958 | 0.18 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr10_-_3827371 | 0.15 |
ENST00000469435.1
|
KLF6
|
Kruppel-like factor 6 |
| chr1_-_23751189 | 0.15 |
ENST00000374601.3
ENST00000450454.2 |
TCEA3
|
transcription elongation factor A (SII), 3 |
| chr12_-_371994 | 0.15 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr3_+_138067314 | 0.15 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr22_-_51017084 | 0.15 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr22_-_51016846 | 0.14 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr1_+_156095951 | 0.14 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr12_+_52430894 | 0.13 |
ENST00000546842.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr19_-_16008880 | 0.13 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr14_-_21493649 | 0.13 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr9_+_97766409 | 0.13 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr19_-_49926698 | 0.13 |
ENST00000270631.1
|
PTH2
|
parathyroid hormone 2 |
| chr2_-_190044480 | 0.13 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr14_-_21493884 | 0.13 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr3_+_138067666 | 0.13 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr2_-_145188137 | 0.13 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_86564696 | 0.12 |
ENST00000437769.1
|
REEP1
|
receptor accessory protein 1 |
| chr5_-_150521192 | 0.11 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr7_+_133615169 | 0.11 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr18_+_12407895 | 0.11 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr1_-_156460391 | 0.11 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr9_-_85882145 | 0.11 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr1_+_22778337 | 0.10 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr12_-_49393092 | 0.10 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr14_-_102704783 | 0.10 |
ENST00000522534.1
|
MOK
|
MOK protein kinase |
| chr22_-_30642782 | 0.10 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr16_-_56223480 | 0.10 |
ENST00000565155.1
|
RP11-461O7.1
|
RP11-461O7.1 |
| chr16_+_50313426 | 0.09 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr2_+_233497931 | 0.09 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr17_-_42188598 | 0.09 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chrX_-_131353461 | 0.09 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr2_-_208031542 | 0.09 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_-_120189900 | 0.09 |
ENST00000546026.1
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chrX_-_15332665 | 0.09 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr1_+_212782012 | 0.09 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr9_+_97766469 | 0.08 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr19_+_16607122 | 0.08 |
ENST00000221671.3
ENST00000594035.1 ENST00000599550.1 ENST00000594813.1 |
C19orf44
|
chromosome 19 open reading frame 44 |
| chr17_-_1389228 | 0.08 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr3_-_137834436 | 0.08 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr1_-_59249732 | 0.08 |
ENST00000371222.2
|
JUN
|
jun proto-oncogene |
| chr11_+_71934962 | 0.08 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr1_+_74701062 | 0.08 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr17_-_42200958 | 0.08 |
ENST00000336057.5
|
HDAC5
|
histone deacetylase 5 |
| chr1_-_19229014 | 0.08 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr10_+_102758105 | 0.08 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr14_-_21493123 | 0.08 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr17_-_1389419 | 0.07 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr1_+_156096336 | 0.07 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr3_-_48598547 | 0.07 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr14_-_22005343 | 0.07 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr17_-_34417479 | 0.07 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr6_+_42984723 | 0.07 |
ENST00000332245.8
|
KLHDC3
|
kelch domain containing 3 |
| chr9_+_116327326 | 0.07 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr20_+_10199566 | 0.06 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr2_-_220117867 | 0.06 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr8_+_1993152 | 0.06 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr11_+_47293795 | 0.06 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr9_-_102582155 | 0.06 |
ENST00000427039.1
|
RP11-554F20.1
|
RP11-554F20.1 |
| chr7_+_75931861 | 0.06 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr7_+_18536090 | 0.06 |
ENST00000441986.1
|
HDAC9
|
histone deacetylase 9 |
| chr6_-_169654139 | 0.06 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr21_-_33984456 | 0.06 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr1_+_77333117 | 0.06 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr5_-_42812143 | 0.06 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_-_204329013 | 0.06 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr4_+_41362615 | 0.05 |
ENST00000509638.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_-_40324767 | 0.05 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr3_+_8543533 | 0.05 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr1_+_84630367 | 0.05 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_+_49057876 | 0.05 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr17_-_42200996 | 0.05 |
ENST00000587135.1
ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5
|
histone deacetylase 5 |
| chr9_-_16728161 | 0.05 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr2_+_33661382 | 0.05 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr16_-_67867749 | 0.05 |
ENST00000566758.1
ENST00000445712.2 ENST00000219172.3 ENST00000564817.1 |
CENPT
|
centromere protein T |
| chr11_-_47374246 | 0.05 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr12_+_15699286 | 0.05 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr7_-_55583740 | 0.05 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr4_+_160203650 | 0.05 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr11_+_7597639 | 0.04 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr3_-_52486841 | 0.04 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr17_+_80186908 | 0.04 |
ENST00000582743.1
ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr8_-_72268889 | 0.04 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr10_-_96829246 | 0.04 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr3_+_8543561 | 0.04 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr17_-_48785216 | 0.04 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr10_-_3827417 | 0.04 |
ENST00000497571.1
ENST00000542957.1 |
KLF6
|
Kruppel-like factor 6 |
| chr11_+_7598239 | 0.04 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr8_+_1993173 | 0.04 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr6_-_137314371 | 0.04 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr14_-_70546897 | 0.04 |
ENST00000394330.2
ENST00000533541.1 ENST00000216568.7 |
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr12_+_25205568 | 0.04 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr22_+_32340447 | 0.03 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr6_+_152128371 | 0.03 |
ENST00000443427.1
|
ESR1
|
estrogen receptor 1 |
| chr20_+_32581525 | 0.03 |
ENST00000246194.3
ENST00000413297.1 |
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chrX_+_70521584 | 0.03 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr8_-_62602327 | 0.03 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr12_-_111358372 | 0.03 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr6_+_44194762 | 0.03 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr12_+_52445191 | 0.03 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr3_+_188664988 | 0.03 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chrX_-_142722897 | 0.03 |
ENST00000338017.4
|
SLITRK4
|
SLIT and NTRK-like family, member 4 |
| chr22_+_32340481 | 0.03 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr12_+_25205446 | 0.03 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr11_-_18034701 | 0.03 |
ENST00000265965.5
|
SERGEF
|
secretion regulating guanine nucleotide exchange factor |
| chr3_+_138067521 | 0.03 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr10_-_97200772 | 0.03 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr6_-_33160231 | 0.03 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr3_-_176914191 | 0.03 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr20_-_62582475 | 0.03 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr14_-_103989033 | 0.03 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chr3_+_54157480 | 0.03 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr10_+_118349920 | 0.02 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr20_+_32581738 | 0.02 |
ENST00000333552.5
|
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr7_-_37488547 | 0.02 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_-_157198860 | 0.02 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr4_+_41362796 | 0.02 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr15_+_68346501 | 0.02 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr11_-_88796803 | 0.02 |
ENST00000418177.2
ENST00000455756.2 |
GRM5
|
glutamate receptor, metabotropic 5 |
| chr5_-_42811986 | 0.02 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr9_-_74980113 | 0.02 |
ENST00000376962.5
ENST00000376960.4 ENST00000237937.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr3_+_35683651 | 0.02 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_+_25205666 | 0.02 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr14_+_76776957 | 0.02 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr1_-_206945830 | 0.02 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr3_+_8543393 | 0.02 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr1_+_145524891 | 0.02 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr10_-_75415825 | 0.02 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr10_+_71561630 | 0.02 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr7_-_15726296 | 0.02 |
ENST00000262041.5
|
MEOX2
|
mesenchyme homeobox 2 |
| chr12_+_52431016 | 0.02 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_-_31146615 | 0.02 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr16_-_88717482 | 0.01 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr8_+_11351494 | 0.01 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr19_-_40324255 | 0.01 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr14_+_101359265 | 0.01 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr3_+_127634312 | 0.01 |
ENST00000407609.3
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr20_+_44657845 | 0.01 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr7_-_37488834 | 0.01 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr20_+_61147651 | 0.01 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr8_+_11351876 | 0.01 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr15_+_42651691 | 0.01 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr7_+_18535893 | 0.01 |
ENST00000432645.2
ENST00000441542.2 |
HDAC9
|
histone deacetylase 9 |
| chr9_-_23779367 | 0.00 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr11_+_1860832 | 0.00 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr7_-_37488777 | 0.00 |
ENST00000445322.1
ENST00000448602.1 |
ELMO1
|
engulfment and cell motility 1 |
| chr8_-_72268968 | 0.00 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr3_+_127634069 | 0.00 |
ENST00000405109.1
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr20_+_44657807 | 0.00 |
ENST00000372315.1
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr8_-_72268721 | 0.00 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr15_+_40226304 | 0.00 |
ENST00000559624.1
ENST00000382727.2 ENST00000263791.5 ENST00000560648.1 |
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4 |
| chr10_+_71561704 | 0.00 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chrX_-_63450480 | 0.00 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr12_-_131200810 | 0.00 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chrX_-_11284095 | 0.00 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |