Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
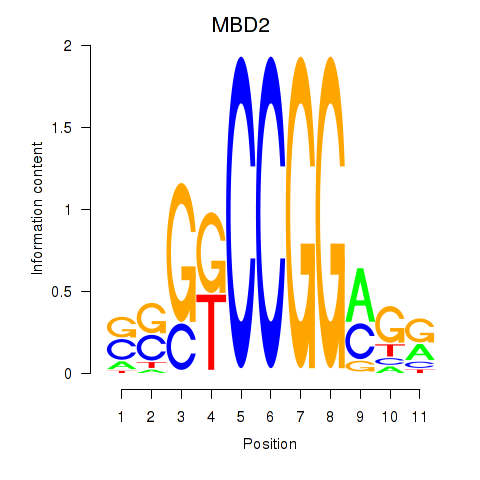
Results for MBD2
Z-value: 1.35
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.7 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MBD2 | hg19_v2_chr18_-_51751132_51751158 | -0.72 | 1.1e-01 | Click! |
Activity profile of MBD2 motif
Sorted Z-values of MBD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_74212073 | 3.83 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr17_-_31404 | 1.09 |
ENST00000343572.7
|
DOC2B
|
double C2-like domains, beta |
| chr1_+_156698743 | 1.00 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr8_-_145688231 | 0.89 |
ENST00000530374.1
|
CYHR1
|
cysteine/histidine-rich 1 |
| chr8_+_61592073 | 0.88 |
ENST00000526846.1
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr11_+_76092353 | 0.87 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr2_+_239756671 | 0.80 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr2_+_42275153 | 0.77 |
ENST00000294964.5
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr6_+_41514305 | 0.77 |
ENST00000409208.1
ENST00000373057.3 |
FOXP4
|
forkhead box P4 |
| chr16_+_2059872 | 0.75 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr5_+_14143728 | 0.73 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr14_-_21566731 | 0.71 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr19_-_14201507 | 0.70 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr19_-_14201776 | 0.69 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr17_+_27920486 | 0.65 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr19_-_41222775 | 0.65 |
ENST00000324464.3
ENST00000450541.1 ENST00000594720.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr5_+_149865838 | 0.64 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr16_-_2059748 | 0.63 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr11_+_118978045 | 0.62 |
ENST00000336702.3
|
C2CD2L
|
C2CD2-like |
| chr16_+_2022036 | 0.61 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr16_-_2059797 | 0.61 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr16_+_57769635 | 0.61 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr4_-_6474173 | 0.60 |
ENST00000382599.4
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr20_+_36531544 | 0.58 |
ENST00000448944.1
|
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr14_-_74416829 | 0.56 |
ENST00000534936.1
|
FAM161B
|
family with sequence similarity 161, member B |
| chr12_+_122516626 | 0.54 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr11_-_64684672 | 0.53 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr19_-_36545649 | 0.53 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr1_+_156698708 | 0.52 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr19_+_36545833 | 0.52 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr19_+_56152361 | 0.50 |
ENST00000585995.1
ENST00000592996.1 |
ZNF581
CCDC106
|
zinc finger protein 581 coiled-coil domain containing 106 |
| chr1_+_43148059 | 0.50 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr19_+_38879061 | 0.49 |
ENST00000587013.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr3_+_14989076 | 0.49 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_+_156698234 | 0.49 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr10_-_79686284 | 0.48 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr17_+_36861735 | 0.44 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr20_-_56284816 | 0.44 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr18_+_19750894 | 0.44 |
ENST00000581694.1
|
GATA6
|
GATA binding protein 6 |
| chr9_-_130616915 | 0.44 |
ENST00000344849.3
|
ENG
|
endoglin |
| chr11_-_47198380 | 0.42 |
ENST00000419701.2
ENST00000526342.1 ENST00000528444.1 ENST00000530596.1 ENST00000525398.1 ENST00000319543.6 ENST00000426335.2 ENST00000527927.1 ENST00000525314.1 |
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr15_-_82338460 | 0.42 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr2_+_220306238 | 0.42 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr5_+_176853702 | 0.42 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr5_+_176853669 | 0.42 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr11_-_75236867 | 0.42 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr3_+_112930946 | 0.42 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr12_-_50222187 | 0.42 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr3_+_33155444 | 0.41 |
ENST00000320954.6
|
CRTAP
|
cartilage associated protein |
| chr19_+_10527449 | 0.41 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr8_+_145137489 | 0.40 |
ENST00000355091.4
ENST00000525087.1 ENST00000361036.6 ENST00000524418.1 |
GPAA1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr5_-_176778803 | 0.39 |
ENST00000303127.7
|
LMAN2
|
lectin, mannose-binding 2 |
| chr10_+_115803650 | 0.39 |
ENST00000369295.2
|
ADRB1
|
adrenoceptor beta 1 |
| chr8_+_144349606 | 0.39 |
ENST00000521682.1
ENST00000340042.1 |
GLI4
|
GLI family zinc finger 4 |
| chr11_-_130786333 | 0.39 |
ENST00000533214.1
ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19
|
sorting nexin 19 |
| chr14_-_74417096 | 0.38 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr1_+_156252708 | 0.38 |
ENST00000295694.5
ENST00000357501.2 |
TMEM79
|
transmembrane protein 79 |
| chr9_-_130617029 | 0.38 |
ENST00000373203.4
|
ENG
|
endoglin |
| chr6_-_31763276 | 0.38 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr20_-_48532046 | 0.38 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr17_+_64961026 | 0.38 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr12_+_120972147 | 0.38 |
ENST00000325954.4
ENST00000542438.1 |
RNF10
|
ring finger protein 10 |
| chr12_+_120972606 | 0.37 |
ENST00000413266.2
|
RNF10
|
ring finger protein 10 |
| chr10_-_123357910 | 0.37 |
ENST00000336553.6
ENST00000457416.2 ENST00000360144.3 ENST00000369059.1 ENST00000356226.4 ENST00000351936.6 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr2_+_11052054 | 0.37 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr1_+_43855560 | 0.37 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr15_-_42783303 | 0.37 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr8_-_141645645 | 0.37 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr5_-_176778523 | 0.36 |
ENST00000513877.1
ENST00000515209.1 ENST00000514458.1 ENST00000502560.1 |
LMAN2
|
lectin, mannose-binding 2 |
| chr14_-_77787198 | 0.36 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr9_-_136933615 | 0.36 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr1_-_179851611 | 0.36 |
ENST00000610272.1
|
RP11-533E19.7
|
RP11-533E19.7 |
| chr16_+_2732476 | 0.36 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr15_-_65715401 | 0.35 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr17_-_80231557 | 0.35 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr1_-_156252590 | 0.35 |
ENST00000361813.5
ENST00000368267.5 |
SMG5
|
SMG5 nonsense mediated mRNA decay factor |
| chr17_-_61920280 | 0.35 |
ENST00000448276.2
ENST00000577990.1 |
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr1_-_27693349 | 0.35 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr1_+_110881945 | 0.34 |
ENST00000602849.1
ENST00000487146.2 |
RBM15
|
RNA binding motif protein 15 |
| chr20_-_48532019 | 0.34 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr7_-_150864635 | 0.34 |
ENST00000297537.4
|
GBX1
|
gastrulation brain homeobox 1 |
| chr9_-_136933092 | 0.33 |
ENST00000357885.2
|
BRD3
|
bromodomain containing 3 |
| chr22_-_19419205 | 0.33 |
ENST00000340170.4
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr2_-_46769694 | 0.33 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr19_-_41196458 | 0.33 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr19_-_16653226 | 0.32 |
ENST00000198939.6
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr1_+_46269248 | 0.32 |
ENST00000361297.2
ENST00000372009.2 |
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr19_+_38397839 | 0.32 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr11_-_46615498 | 0.32 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr12_+_6875519 | 0.32 |
ENST00000389462.4
ENST00000540874.1 ENST00000309083.6 |
PTMS
|
parathymosin |
| chr14_-_99737822 | 0.32 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr12_+_68042517 | 0.32 |
ENST00000393555.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr17_-_79876010 | 0.31 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr12_+_7037461 | 0.31 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr11_-_67141090 | 0.31 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr3_-_183979251 | 0.31 |
ENST00000296238.3
|
CAMK2N2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr5_+_170846640 | 0.31 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr16_+_3019246 | 0.31 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr2_-_208031542 | 0.31 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_-_156698591 | 0.31 |
ENST00000368219.1
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr22_-_20792089 | 0.31 |
ENST00000405555.3
ENST00000266214.5 |
SCARF2
|
scavenger receptor class F, member 2 |
| chr19_-_41196534 | 0.31 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr19_-_42759300 | 0.31 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr2_+_181845532 | 0.30 |
ENST00000602475.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr19_+_56152262 | 0.30 |
ENST00000325333.5
ENST00000590190.1 |
ZNF580
|
zinc finger protein 580 |
| chr14_-_93581615 | 0.30 |
ENST00000555553.1
ENST00000555495.1 ENST00000554999.1 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr14_+_102829300 | 0.30 |
ENST00000359520.7
|
TECPR2
|
tectonin beta-propeller repeat containing 2 |
| chr10_-_105615164 | 0.30 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr14_-_103523745 | 0.30 |
ENST00000361246.2
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr1_+_153606532 | 0.30 |
ENST00000403433.1
|
CHTOP
|
chromatin target of PRMT1 |
| chr14_+_77787227 | 0.30 |
ENST00000216465.5
ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr11_+_65383227 | 0.30 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr5_+_153570319 | 0.30 |
ENST00000377661.2
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr19_+_42388437 | 0.29 |
ENST00000378152.4
ENST00000337665.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr19_+_14544099 | 0.29 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr11_-_62414070 | 0.29 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr5_-_156569850 | 0.29 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr8_+_37654424 | 0.29 |
ENST00000315215.7
|
GPR124
|
G protein-coupled receptor 124 |
| chr16_-_30905584 | 0.29 |
ENST00000380317.4
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr6_-_10415218 | 0.29 |
ENST00000466073.1
ENST00000498450.1 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr17_-_61919812 | 0.29 |
ENST00000225742.9
|
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr17_+_46125685 | 0.29 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr10_+_104404218 | 0.29 |
ENST00000302424.7
|
TRIM8
|
tripartite motif containing 8 |
| chr19_-_4124079 | 0.29 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr1_-_153919128 | 0.29 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr11_-_64851496 | 0.28 |
ENST00000404147.3
ENST00000275517.3 |
CDCA5
|
cell division cycle associated 5 |
| chr1_-_32229523 | 0.28 |
ENST00000398547.1
ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr11_-_67169265 | 0.28 |
ENST00000358239.4
ENST00000376745.4 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr5_+_153570285 | 0.28 |
ENST00000425427.2
ENST00000297107.6 |
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr19_+_36208877 | 0.28 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr17_-_66097610 | 0.28 |
ENST00000584047.1
ENST00000579629.1 |
AC145343.2
|
AC145343.2 |
| chr14_+_21538517 | 0.28 |
ENST00000298693.3
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr2_+_241375069 | 0.28 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr16_+_3019552 | 0.28 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr3_-_53080047 | 0.27 |
ENST00000482396.1
ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr4_+_2061119 | 0.27 |
ENST00000423729.2
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative) |
| chr16_+_3019309 | 0.27 |
ENST00000576565.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr5_-_54523143 | 0.27 |
ENST00000513312.1
|
MCIDAS
|
multiciliate differentiation and DNA synthesis associated cell cycle protein |
| chr17_-_5372229 | 0.27 |
ENST00000433302.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr16_-_51185149 | 0.27 |
ENST00000566102.1
ENST00000541611.1 |
SALL1
|
spalt-like transcription factor 1 |
| chr3_+_33155525 | 0.27 |
ENST00000449224.1
|
CRTAP
|
cartilage associated protein |
| chr6_-_16761678 | 0.27 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr11_-_67169253 | 0.27 |
ENST00000527663.1
ENST00000312989.7 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr10_-_103880209 | 0.27 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr7_+_129710350 | 0.26 |
ENST00000335420.5
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr5_+_133861790 | 0.26 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr6_+_41514078 | 0.26 |
ENST00000373063.3
ENST00000373060.1 |
FOXP4
|
forkhead box P4 |
| chr2_-_119605253 | 0.26 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr11_-_6633799 | 0.26 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr11_+_61735266 | 0.26 |
ENST00000601917.1
|
AP003733.1
|
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
| chr9_+_124413873 | 0.26 |
ENST00000408936.3
|
DAB2IP
|
DAB2 interacting protein |
| chr11_-_64545222 | 0.26 |
ENST00000433274.2
ENST00000432725.1 |
SF1
|
splicing factor 1 |
| chr13_+_98795505 | 0.26 |
ENST00000319562.6
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr3_+_71803201 | 0.26 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr11_+_46354455 | 0.26 |
ENST00000343674.6
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr1_+_156084461 | 0.26 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr20_-_1447547 | 0.26 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr12_-_120554622 | 0.26 |
ENST00000229340.5
|
RAB35
|
RAB35, member RAS oncogene family |
| chr16_-_22385901 | 0.26 |
ENST00000268383.2
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa |
| chr9_-_136933134 | 0.25 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr19_-_1568057 | 0.25 |
ENST00000402693.4
ENST00000388824.6 |
MEX3D
|
mex-3 RNA binding family member D |
| chr6_+_148663729 | 0.25 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr19_-_16653325 | 0.25 |
ENST00000546361.2
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr1_+_179851893 | 0.25 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr9_-_35732362 | 0.25 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chr12_+_57482877 | 0.25 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr2_-_219537134 | 0.25 |
ENST00000295704.2
|
RNF25
|
ring finger protein 25 |
| chr20_-_1447467 | 0.25 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr7_+_69064566 | 0.25 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr1_+_153606513 | 0.25 |
ENST00000368694.3
|
CHTOP
|
chromatin target of PRMT1 |
| chr1_-_43232649 | 0.25 |
ENST00000372526.2
ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr17_-_27277615 | 0.25 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr20_-_46415297 | 0.25 |
ENST00000467815.1
ENST00000359930.4 |
SULF2
|
sulfatase 2 |
| chr15_+_40733387 | 0.25 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr11_-_67271723 | 0.24 |
ENST00000533391.1
ENST00000534749.1 ENST00000532703.1 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr12_+_110152033 | 0.24 |
ENST00000538780.1
|
FAM222A
|
family with sequence similarity 222, member A |
| chr6_+_1389989 | 0.24 |
ENST00000259806.1
|
FOXF2
|
forkhead box F2 |
| chr5_+_179159813 | 0.24 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr5_-_72744336 | 0.24 |
ENST00000499003.3
|
FOXD1
|
forkhead box D1 |
| chr19_+_36545781 | 0.24 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr1_-_156698181 | 0.24 |
ENST00000313146.6
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr16_-_4166186 | 0.24 |
ENST00000294016.3
|
ADCY9
|
adenylate cyclase 9 |
| chr12_-_120554534 | 0.24 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr7_+_69064300 | 0.24 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr1_-_40237020 | 0.24 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr1_+_27561007 | 0.24 |
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr2_+_242641442 | 0.24 |
ENST00000313552.6
ENST00000406941.1 |
ING5
|
inhibitor of growth family, member 5 |
| chr11_+_67056755 | 0.24 |
ENST00000511455.2
ENST00000308440.6 |
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr14_+_21538429 | 0.24 |
ENST00000298694.4
ENST00000555038.1 |
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr17_+_48133459 | 0.24 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr11_+_67055986 | 0.23 |
ENST00000447274.2
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr17_-_48227877 | 0.23 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr19_+_18496957 | 0.23 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr1_-_23857698 | 0.23 |
ENST00000361729.2
|
E2F2
|
E2F transcription factor 2 |
| chr14_+_53019822 | 0.23 |
ENST00000321662.6
|
GPR137C
|
G protein-coupled receptor 137C |
| chr6_-_43596899 | 0.23 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr19_-_4066890 | 0.23 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr7_-_19157248 | 0.23 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr3_+_49591881 | 0.23 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr13_+_98795434 | 0.22 |
ENST00000376586.2
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
Network of associatons between targets according to the STRING database.
First level regulatory network of MBD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 0.8 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.2 | 0.2 | GO:0060922 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.2 | 0.7 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.2 | 0.5 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.2 | 0.6 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.2 | 0.2 | GO:0060932 | bundle of His development(GO:0003166) His-Purkinje system cell differentiation(GO:0060932) |
| 0.2 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.4 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.4 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.1 | 0.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.9 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.7 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.6 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.5 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.1 | 0.2 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) |
| 0.1 | 0.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.3 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.1 | 0.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.3 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.5 | GO:0021622 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.5 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.3 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.4 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 2.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.8 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.7 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.4 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 0.4 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.6 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.4 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.2 | GO:0014040 | acetaldehyde metabolic process(GO:0006117) positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.1 | 0.2 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 1.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.6 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.2 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.3 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.4 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.4 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.3 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0009093 | cysteine biosynthetic process from serine(GO:0006535) cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0048003 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0061184 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.3 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.0 | 0.2 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.6 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.8 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.6 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.4 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.1 | GO:0060684 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.2 | 0.6 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.7 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.2 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 0.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 1.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 2.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 0.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.4 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.4 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.3 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.1 | 0.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.6 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.5 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0050421 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |