Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
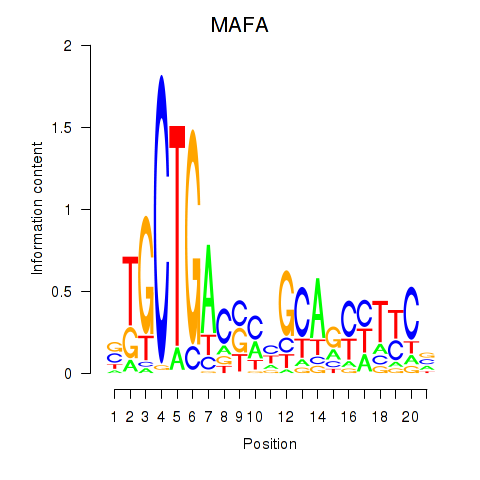
Results for MAFA
Z-value: 0.70
Transcription factors associated with MAFA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFA
|
ENSG00000182759.3 | MAF bZIP transcription factor A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFA | hg19_v2_chr8_-_144512576_144512576 | -0.89 | 1.7e-02 | Click! |
Activity profile of MAFA motif
Sorted Z-values of MAFA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_30951282 | 0.82 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr9_+_100000717 | 0.38 |
ENST00000375205.2
ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180
|
coiled-coil domain containing 180 |
| chr9_-_130712995 | 0.35 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr16_-_21436459 | 0.34 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr11_+_777562 | 0.33 |
ENST00000530083.1
|
AP006621.5
|
Protein LOC100506518 |
| chr11_+_113930955 | 0.29 |
ENST00000535700.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr10_-_79397391 | 0.27 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_+_107461948 | 0.24 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr5_-_179285785 | 0.23 |
ENST00000520698.1
ENST00000518235.1 ENST00000376931.2 ENST00000518219.1 ENST00000521333.1 ENST00000523084.1 |
C5orf45
|
chromosome 5 open reading frame 45 |
| chr11_-_777467 | 0.22 |
ENST00000397472.2
ENST00000524550.1 ENST00000319863.8 ENST00000526325.1 ENST00000442059.2 |
PDDC1
|
Parkinson disease 7 domain containing 1 |
| chr16_-_67514982 | 0.22 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr10_-_118897567 | 0.21 |
ENST00000369206.5
|
VAX1
|
ventral anterior homeobox 1 |
| chr10_-_29811456 | 0.20 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr1_+_110754094 | 0.20 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr7_-_150754935 | 0.20 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr19_+_38880695 | 0.19 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_-_179285848 | 0.18 |
ENST00000403396.2
ENST00000292586.6 |
C5orf45
|
chromosome 5 open reading frame 45 |
| chr1_-_45956822 | 0.18 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr16_+_22524844 | 0.18 |
ENST00000538606.1
ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr21_+_44589118 | 0.18 |
ENST00000291554.2
|
CRYAA
|
crystallin, alpha A |
| chr6_-_24489842 | 0.17 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chrX_-_47509994 | 0.17 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chr17_-_3796334 | 0.17 |
ENST00000381771.2
ENST00000348335.2 ENST00000158166.5 |
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr3_+_52422899 | 0.17 |
ENST00000480649.1
|
DNAH1
|
dynein, axonemal, heavy chain 1 |
| chr3_+_42190714 | 0.16 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr10_-_49701686 | 0.16 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr1_+_221054584 | 0.15 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr16_-_29415350 | 0.15 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr17_-_17109579 | 0.15 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr12_-_22487588 | 0.15 |
ENST00000381424.3
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr10_-_102289611 | 0.14 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr7_-_86849836 | 0.14 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr8_+_145438870 | 0.14 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr12_-_123215306 | 0.14 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr7_-_135412925 | 0.14 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr16_-_29517141 | 0.14 |
ENST00000550665.1
|
RP11-231C14.4
|
Uncharacterized protein |
| chr5_+_176853702 | 0.14 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr7_-_155160629 | 0.14 |
ENST00000378120.5
|
BLACE
|
B-cell acute lymphoblastic leukemia expressed |
| chr12_+_120137573 | 0.13 |
ENST00000535109.1
|
RP1-127H14.3
|
Uncharacterized protein |
| chr11_+_72929319 | 0.13 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr2_+_102624977 | 0.13 |
ENST00000441002.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chrX_-_153141783 | 0.12 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr1_-_45956868 | 0.12 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr20_+_306177 | 0.12 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr3_+_184016986 | 0.11 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr11_+_72929402 | 0.11 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr20_+_306221 | 0.11 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr16_-_87525651 | 0.11 |
ENST00000268616.4
|
ZCCHC14
|
zinc finger, CCHC domain containing 14 |
| chr7_+_100860949 | 0.11 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr9_+_131549610 | 0.11 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr12_-_22487618 | 0.11 |
ENST00000404299.3
ENST00000396037.4 |
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr1_+_221054411 | 0.10 |
ENST00000427693.1
|
HLX
|
H2.0-like homeobox |
| chr7_-_73133959 | 0.10 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr16_-_46865047 | 0.10 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr10_-_75676400 | 0.10 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr19_-_50464338 | 0.09 |
ENST00000426971.2
ENST00000447370.2 |
SIGLEC11
|
sialic acid binding Ig-like lectin 11 |
| chr1_-_45956800 | 0.09 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr14_+_50779071 | 0.09 |
ENST00000426751.2
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chrX_-_47509887 | 0.09 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr12_+_53817633 | 0.09 |
ENST00000257863.4
ENST00000550311.1 ENST00000379791.3 |
AMHR2
|
anti-Mullerian hormone receptor, type II |
| chr15_-_41047421 | 0.09 |
ENST00000560460.1
ENST00000338376.3 ENST00000560905.1 |
RMDN3
|
regulator of microtubule dynamics 3 |
| chrX_+_68725084 | 0.09 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr11_-_70507901 | 0.09 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr18_-_2982869 | 0.09 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr2_-_26205550 | 0.09 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
| chr16_+_28996572 | 0.09 |
ENST00000360872.5
ENST00000566177.1 ENST00000354453.4 |
LAT
|
linker for activation of T cells |
| chr9_+_97488939 | 0.08 |
ENST00000277198.2
ENST00000297979.5 |
C9orf3
|
chromosome 9 open reading frame 3 |
| chr11_+_118826999 | 0.08 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr22_+_48972118 | 0.08 |
ENST00000358295.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr15_-_90456114 | 0.07 |
ENST00000398333.3
|
C15orf38-AP3S2
|
C15orf38-AP3S2 readthrough |
| chr3_-_49823941 | 0.07 |
ENST00000321599.4
ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1
|
inositol hexakisphosphate kinase 1 |
| chr4_-_10118348 | 0.07 |
ENST00000502702.1
|
WDR1
|
WD repeat domain 1 |
| chr19_+_21541732 | 0.07 |
ENST00000311015.3
|
ZNF738
|
zinc finger protein 738 |
| chr7_-_100860851 | 0.07 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr6_-_143832793 | 0.07 |
ENST00000438118.2
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr9_-_100000957 | 0.07 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr19_+_36103631 | 0.06 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr3_+_196439170 | 0.06 |
ENST00000392391.3
ENST00000314118.4 |
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr3_+_131100515 | 0.06 |
ENST00000537561.1
ENST00000359850.3 ENST00000521288.1 ENST00000502852.1 |
NUDT16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr15_-_23086259 | 0.06 |
ENST00000538684.1
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr5_-_22853429 | 0.06 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr19_+_21541777 | 0.06 |
ENST00000380870.4
ENST00000597810.1 ENST00000594245.1 |
ZNF738
|
zinc finger protein 738 |
| chrX_-_153141302 | 0.06 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr19_-_2050852 | 0.06 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr19_+_18451391 | 0.06 |
ENST00000269919.6
ENST00000604499.2 ENST00000595066.1 ENST00000252813.5 |
PGPEP1
|
pyroglutamyl-peptidase I |
| chr20_-_30795511 | 0.06 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chr19_-_4124079 | 0.05 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr9_-_28670283 | 0.05 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr5_+_138629628 | 0.05 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr10_+_135122906 | 0.05 |
ENST00000368554.4
|
ZNF511
|
zinc finger protein 511 |
| chr15_+_98503922 | 0.05 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr16_-_21868739 | 0.05 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr15_-_90456156 | 0.05 |
ENST00000357484.5
|
C15orf38
|
chromosome 15 open reading frame 38 |
| chr10_-_135171178 | 0.05 |
ENST00000368551.1
|
FUOM
|
fucose mutarotase |
| chr4_+_72052964 | 0.05 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr19_+_33182823 | 0.05 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr1_+_174769006 | 0.05 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr6_+_22221010 | 0.05 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr10_-_79397479 | 0.04 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_+_56546363 | 0.04 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr15_+_80696666 | 0.04 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_+_90286562 | 0.04 |
ENST00000525774.1
ENST00000337338.5 |
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr8_-_63998590 | 0.04 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr9_-_130889990 | 0.04 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr16_+_28996364 | 0.04 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr2_+_191745560 | 0.04 |
ENST00000338435.4
|
GLS
|
glutaminase |
| chr22_-_28197486 | 0.04 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr4_+_72053017 | 0.04 |
ENST00000351898.6
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr16_+_70258261 | 0.04 |
ENST00000594734.1
|
FKSG63
|
FKSG63 |
| chr6_+_42896865 | 0.04 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr11_-_75380165 | 0.04 |
ENST00000304771.3
|
MAP6
|
microtubule-associated protein 6 |
| chrX_-_1511617 | 0.03 |
ENST00000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr18_+_77623668 | 0.03 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr11_-_57518848 | 0.03 |
ENST00000436147.3
|
BTBD18
|
BTB (POZ) domain containing 18 |
| chr5_-_94620239 | 0.03 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr7_-_86849883 | 0.03 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr19_+_39647271 | 0.03 |
ENST00000599657.1
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr15_-_23086394 | 0.03 |
ENST00000337435.4
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr1_+_163039143 | 0.03 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr7_-_30029328 | 0.03 |
ENST00000425819.2
ENST00000409570.1 |
SCRN1
|
secernin 1 |
| chr4_-_151936416 | 0.03 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr10_+_124221036 | 0.03 |
ENST00000368984.3
|
HTRA1
|
HtrA serine peptidase 1 |
| chr14_+_50779029 | 0.02 |
ENST00000245448.6
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr6_+_7727030 | 0.02 |
ENST00000283147.6
|
BMP6
|
bone morphogenetic protein 6 |
| chr1_+_200842083 | 0.02 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chr1_-_54872059 | 0.02 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr4_+_15005391 | 0.02 |
ENST00000507071.1
ENST00000345451.3 ENST00000259997.5 ENST00000382395.3 ENST00000382401.3 |
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr19_+_18451439 | 0.02 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr2_+_103236004 | 0.02 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr3_+_140770183 | 0.02 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr5_+_60933634 | 0.02 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr10_-_135171510 | 0.02 |
ENST00000278025.4
ENST00000368552.3 |
FUOM
|
fucose mutarotase |
| chr17_-_47925379 | 0.02 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr10_+_102505468 | 0.02 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chr2_+_191745535 | 0.02 |
ENST00000320717.3
|
GLS
|
glutaminase |
| chr13_+_28194873 | 0.02 |
ENST00000302979.3
|
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa |
| chr19_+_12949251 | 0.01 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chrY_-_1461617 | 0.01 |
ENSTR0000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr8_-_114389353 | 0.01 |
ENST00000343508.3
|
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr4_-_151936865 | 0.01 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr3_-_48754599 | 0.01 |
ENST00000413654.1
ENST00000454335.1 ENST00000440424.1 ENST00000449610.1 ENST00000443964.1 ENST00000417896.1 ENST00000413298.1 ENST00000449563.1 ENST00000443853.1 ENST00000437427.1 ENST00000446860.1 ENST00000412850.1 ENST00000424035.1 ENST00000340879.4 ENST00000431721.2 ENST00000434860.1 ENST00000328631.5 ENST00000432678.2 |
IP6K2
|
inositol hexakisphosphate kinase 2 |
| chr3_+_185304059 | 0.01 |
ENST00000427465.2
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chrX_+_74493896 | 0.01 |
ENST00000373383.4
ENST00000373379.1 |
UPRT
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr7_-_151433342 | 0.01 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_+_107462471 | 0.01 |
ENST00000600612.1
|
AP000889.3
|
HCG2032453; Uncharacterized protein; cDNA FLJ25337 fis, clone TST00714 |
| chr6_-_139613269 | 0.01 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chrX_-_53310791 | 0.01 |
ENST00000375365.2
|
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr5_+_138629337 | 0.01 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr6_-_99797522 | 0.01 |
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila) |
| chr17_-_39781054 | 0.00 |
ENST00000463128.1
|
KRT17
|
keratin 17 |
| chr17_+_38465441 | 0.00 |
ENST00000577646.1
ENST00000254066.5 |
RARA
|
retinoic acid receptor, alpha |
| chr17_-_41623009 | 0.00 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr17_-_76128488 | 0.00 |
ENST00000322914.3
|
TMC6
|
transmembrane channel-like 6 |
| chr10_+_102505954 | 0.00 |
ENST00000556085.1
ENST00000427256.1 |
PAX2
|
paired box 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.2 | GO:0010983 | negative regulation of triglyceride catabolic process(GO:0010897) regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.2 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.0 | GO:0052251 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.2 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |