Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
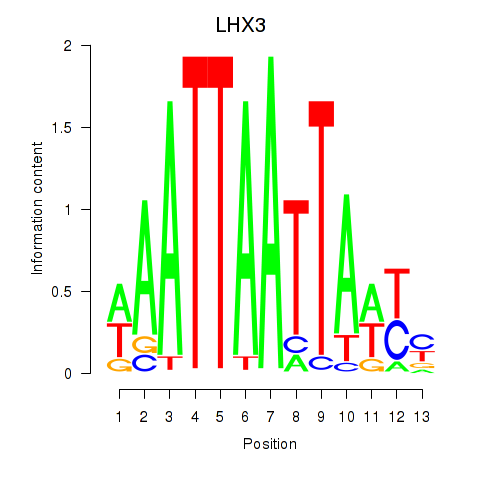
Results for LHX3
Z-value: 0.38
Transcription factors associated with LHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX3
|
ENSG00000107187.11 | LIM homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX3 | hg19_v2_chr9_-_139094988_139095012 | 0.32 | 5.4e-01 | Click! |
Activity profile of LHX3 motif
Sorted Z-values of LHX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_61151306 | 0.36 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr11_-_63376013 | 0.32 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr14_+_29241910 | 0.28 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr14_-_57197224 | 0.27 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr12_-_118628315 | 0.26 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr4_-_120243545 | 0.25 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr8_-_66750978 | 0.25 |
ENST00000523253.1
|
PDE7A
|
phosphodiesterase 7A |
| chr2_+_29001711 | 0.24 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr6_-_138833630 | 0.14 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr18_+_61616510 | 0.13 |
ENST00000408945.3
|
HMSD
|
histocompatibility (minor) serpin domain containing |
| chr15_+_75970672 | 0.12 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr4_+_3344141 | 0.12 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr22_+_23487513 | 0.10 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr14_-_21566731 | 0.10 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr6_-_47445214 | 0.09 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr9_-_21482312 | 0.09 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr17_-_4938712 | 0.09 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr16_+_89334512 | 0.09 |
ENST00000602042.1
|
AC137932.1
|
AC137932.1 |
| chr9_-_128246769 | 0.09 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr6_+_26104104 | 0.08 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr8_-_25281747 | 0.08 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr10_-_90611566 | 0.08 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr11_-_58378759 | 0.08 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr13_+_24144796 | 0.08 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr19_+_48949087 | 0.07 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr19_+_11457232 | 0.07 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr14_-_45252031 | 0.07 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr16_-_28937027 | 0.07 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr17_-_8263538 | 0.07 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr3_+_186692745 | 0.07 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr5_+_59783941 | 0.06 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr4_+_74301880 | 0.06 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chrX_+_107288239 | 0.06 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_-_52761262 | 0.06 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr19_+_48949030 | 0.06 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chrX_+_107288197 | 0.06 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_+_134671234 | 0.06 |
ENST00000436302.2
ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein-like 3 |
| chr8_+_119294456 | 0.06 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr14_-_101295407 | 0.06 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr9_+_125133315 | 0.06 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr19_+_11457175 | 0.06 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr7_-_84122033 | 0.05 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr14_+_55494323 | 0.05 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr17_-_39623681 | 0.05 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chrX_-_13835147 | 0.05 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr1_+_244214577 | 0.05 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr7_+_30185496 | 0.05 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr9_-_100000957 | 0.05 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr1_-_48866517 | 0.05 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr6_+_130339710 | 0.05 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr17_-_72772462 | 0.05 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr12_-_118628350 | 0.05 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr7_+_30185406 | 0.05 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr10_+_695888 | 0.05 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr11_-_559377 | 0.05 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr9_+_125132803 | 0.05 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr3_-_20053741 | 0.04 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr16_+_15489629 | 0.04 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr14_+_55493920 | 0.04 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr18_+_616711 | 0.04 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr1_-_231005310 | 0.04 |
ENST00000470540.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr15_+_64680003 | 0.04 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr2_+_26624775 | 0.04 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr1_+_15668240 | 0.04 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr19_-_51920952 | 0.03 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr14_+_20215587 | 0.03 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr2_-_99871570 | 0.03 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr1_-_94147385 | 0.03 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr15_+_65843130 | 0.03 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr1_-_24126023 | 0.03 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr12_+_49740700 | 0.03 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr17_-_72772425 | 0.03 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr7_-_4901625 | 0.03 |
ENST00000404991.1
|
PAPOLB
|
poly(A) polymerase beta (testis specific) |
| chr18_+_616672 | 0.03 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr10_+_104263743 | 0.03 |
ENST00000369902.3
ENST00000369899.2 ENST00000423559.2 |
SUFU
|
suppressor of fused homolog (Drosophila) |
| chr10_+_18549645 | 0.03 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr5_+_140529630 | 0.03 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr2_+_135596106 | 0.03 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr2_+_54785485 | 0.03 |
ENST00000333896.5
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_+_696000 | 0.03 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr16_+_15489603 | 0.03 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr15_-_75748143 | 0.03 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr14_+_21566980 | 0.02 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr17_-_27418537 | 0.02 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr11_-_128894053 | 0.02 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chrM_+_10053 | 0.02 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr13_-_81801115 | 0.02 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr14_-_21567009 | 0.02 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr7_-_122840015 | 0.02 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr7_-_140482926 | 0.02 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr12_-_86650077 | 0.02 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_57123815 | 0.02 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr16_+_28565230 | 0.02 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr12_-_86650045 | 0.02 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr4_-_185275104 | 0.02 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr10_-_27529486 | 0.02 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr5_+_169011033 | 0.02 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_50016610 | 0.02 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_+_35851570 | 0.01 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr3_-_48130707 | 0.01 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr15_-_99057551 | 0.01 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr4_-_41884620 | 0.01 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr5_-_1882858 | 0.01 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chrX_+_9431324 | 0.01 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr4_-_23891658 | 0.01 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chrX_-_110655306 | 0.01 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr14_+_101295638 | 0.01 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr1_-_24126051 | 0.01 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr19_+_50016411 | 0.01 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_-_49712123 | 0.01 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_9933500 | 0.01 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr4_-_36245561 | 0.01 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_+_71384257 | 0.01 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr16_-_28634874 | 0.01 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr3_-_47517302 | 0.01 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr3_+_148415571 | 0.01 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr10_-_128359074 | 0.01 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr4_+_41937131 | 0.01 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr2_-_145278475 | 0.01 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_-_160924589 | 0.01 |
ENST00000368029.3
|
ITLN2
|
intelectin 2 |
| chr11_-_67980744 | 0.00 |
ENST00000401547.2
ENST00000453170.1 ENST00000304363.4 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr5_+_166711804 | 0.00 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr9_+_125133467 | 0.00 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_+_59989918 | 0.00 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chrX_+_108779004 | 0.00 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr12_+_40787194 | 0.00 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr1_+_168250194 | 0.00 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr19_-_43969796 | 0.00 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr2_-_99279928 | 0.00 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr6_-_49712072 | 0.00 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr9_-_116065551 | 0.00 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr10_-_74283694 | 0.00 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr2_-_207078154 | 0.00 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr1_+_81771806 | 0.00 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr8_-_30670384 | 0.00 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |