Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
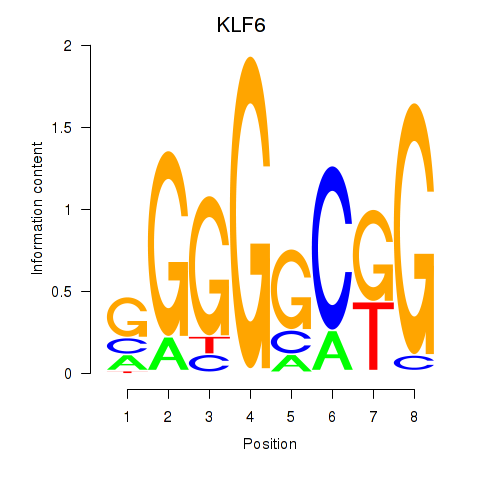
Results for KLF6
Z-value: 0.48
Transcription factors associated with KLF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF6
|
ENSG00000067082.10 | Kruppel like factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF6 | hg19_v2_chr10_-_3827371_3827386 | 0.63 | 1.8e-01 | Click! |
Activity profile of KLF6 motif
Sorted Z-values of KLF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_16435625 | 0.28 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr3_+_50192457 | 0.26 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr20_-_31172598 | 0.26 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr12_-_50222187 | 0.25 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr1_+_156698708 | 0.25 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr22_+_38004832 | 0.24 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr1_+_156698234 | 0.23 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr11_+_48002279 | 0.23 |
ENST00000534219.1
ENST00000527952.1 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr1_+_156698743 | 0.22 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr11_-_65321198 | 0.21 |
ENST00000530426.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr19_-_14117074 | 0.21 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr22_+_38004942 | 0.18 |
ENST00000439161.1
ENST00000449944.1 ENST00000411501.1 ENST00000453208.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr14_+_100705322 | 0.18 |
ENST00000262238.4
|
YY1
|
YY1 transcription factor |
| chr2_+_105471969 | 0.18 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr17_-_73874654 | 0.18 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr17_+_79008940 | 0.17 |
ENST00000392411.3
ENST00000575989.1 ENST00000321280.7 ENST00000428708.2 ENST00000575712.1 ENST00000575245.1 ENST00000435091.3 ENST00000321300.6 |
BAIAP2
|
BAI1-associated protein 2 |
| chr6_+_31588478 | 0.17 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr14_-_23451845 | 0.17 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr6_+_44095263 | 0.17 |
ENST00000532634.1
|
TMEM63B
|
transmembrane protein 63B |
| chr15_-_75743915 | 0.17 |
ENST00000394949.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr19_+_50084561 | 0.17 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr1_-_156698181 | 0.17 |
ENST00000313146.6
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr17_-_42277203 | 0.16 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr1_+_43148059 | 0.16 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr22_+_45680822 | 0.16 |
ENST00000216211.4
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr1_-_156698591 | 0.16 |
ENST00000368219.1
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr11_-_65640071 | 0.15 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr10_+_76586348 | 0.15 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr17_-_42994283 | 0.15 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr15_-_75744014 | 0.15 |
ENST00000394947.3
ENST00000565264.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr1_+_154297988 | 0.15 |
ENST00000368487.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr20_+_48807351 | 0.15 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr17_-_80231557 | 0.15 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr17_-_7145475 | 0.15 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr4_+_183370146 | 0.14 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr22_+_38321840 | 0.14 |
ENST00000454685.1
|
MICALL1
|
MICAL-like 1 |
| chr5_+_141016969 | 0.14 |
ENST00000518856.1
|
RELL2
|
RELT-like 2 |
| chr22_+_38004723 | 0.14 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr9_-_117267717 | 0.14 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr15_+_96873921 | 0.14 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr19_-_49568311 | 0.14 |
ENST00000595857.1
ENST00000451356.2 |
NTF4
|
neurotrophin 4 |
| chr11_-_119234876 | 0.14 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr19_+_45504688 | 0.13 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr15_-_72410455 | 0.13 |
ENST00000569314.1
|
MYO9A
|
myosin IXA |
| chr14_+_75230011 | 0.13 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr1_+_198126209 | 0.13 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr7_+_143078652 | 0.13 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr19_-_47734448 | 0.13 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr11_-_63933504 | 0.12 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr17_+_47572647 | 0.12 |
ENST00000172229.3
|
NGFR
|
nerve growth factor receptor |
| chr3_-_184079382 | 0.12 |
ENST00000344937.7
ENST00000423355.2 ENST00000434054.2 ENST00000457512.1 ENST00000265593.4 |
CLCN2
|
chloride channel, voltage-sensitive 2 |
| chr17_-_27277615 | 0.12 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr17_+_59477233 | 0.12 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr15_-_76352069 | 0.12 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr5_+_42423872 | 0.12 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr3_-_141868293 | 0.12 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr22_+_38004473 | 0.12 |
ENST00000414350.3
ENST00000343632.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr19_-_41220957 | 0.11 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr6_+_99282570 | 0.11 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr7_-_99774945 | 0.11 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr17_-_31404 | 0.11 |
ENST00000343572.7
|
DOC2B
|
double C2-like domains, beta |
| chr17_+_4736812 | 0.11 |
ENST00000453408.3
|
MINK1
|
misshapen-like kinase 1 |
| chr15_+_41136216 | 0.11 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr5_+_149865838 | 0.11 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr7_-_129592471 | 0.10 |
ENST00000473814.2
ENST00000490974.1 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr1_+_26496362 | 0.10 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr19_-_31840438 | 0.10 |
ENST00000240587.4
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr3_+_159481464 | 0.10 |
ENST00000467377.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr3_+_150126101 | 0.10 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chr17_+_70117153 | 0.10 |
ENST00000245479.2
|
SOX9
|
SRY (sex determining region Y)-box 9 |
| chr7_+_148936732 | 0.10 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr3_+_10289707 | 0.10 |
ENST00000287652.4
|
TATDN2
|
TatD DNase domain containing 2 |
| chr8_-_144890847 | 0.10 |
ENST00000531942.1
|
SCRIB
|
scribbled planar cell polarity protein |
| chr9_+_2017063 | 0.10 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_-_115249484 | 0.10 |
ENST00000457681.1
|
C9orf147
|
chromosome 9 open reading frame 147 |
| chr7_-_72936608 | 0.10 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr6_+_1389989 | 0.09 |
ENST00000259806.1
|
FOXF2
|
forkhead box F2 |
| chr19_+_36545833 | 0.09 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr7_-_27213893 | 0.09 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr5_+_170846640 | 0.09 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr14_-_23451467 | 0.09 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr19_+_50270219 | 0.09 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr2_+_42275153 | 0.09 |
ENST00000294964.5
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr17_-_7832753 | 0.09 |
ENST00000303790.2
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr8_-_80680078 | 0.09 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr17_-_74733404 | 0.09 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr11_+_45907177 | 0.09 |
ENST00000241014.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr3_-_53080047 | 0.09 |
ENST00000482396.1
ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr1_+_27022839 | 0.09 |
ENST00000457599.2
|
ARID1A
|
AT rich interactive domain 1A (SWI-like) |
| chr21_+_47743995 | 0.09 |
ENST00000359568.5
|
PCNT
|
pericentrin |
| chr22_+_31608219 | 0.09 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr1_+_40839369 | 0.09 |
ENST00000372718.3
|
SMAP2
|
small ArfGAP2 |
| chr17_-_7297519 | 0.09 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr17_+_4736627 | 0.09 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr4_+_4861385 | 0.09 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr1_+_240255166 | 0.09 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr6_+_31926857 | 0.09 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr11_+_65479462 | 0.09 |
ENST00000377046.3
ENST00000352980.4 ENST00000341318.4 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr17_+_38296576 | 0.09 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
| chr10_+_76585303 | 0.09 |
ENST00000372725.1
|
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr17_-_40273348 | 0.09 |
ENST00000225916.5
|
KAT2A
|
K(lysine) acetyltransferase 2A |
| chr17_-_56065540 | 0.08 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr17_+_7452442 | 0.08 |
ENST00000557233.1
|
TNFSF12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr12_+_862089 | 0.08 |
ENST00000315939.6
ENST00000537687.1 ENST00000447667.2 |
WNK1
|
WNK lysine deficient protein kinase 1 |
| chr1_+_151693984 | 0.08 |
ENST00000479191.1
|
RIIAD1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
| chr2_-_220408430 | 0.08 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr1_-_205649580 | 0.08 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr20_+_306221 | 0.08 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr20_+_306177 | 0.08 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr1_+_160336851 | 0.08 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr19_-_10444188 | 0.08 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr11_+_65480222 | 0.08 |
ENST00000534681.1
|
KAT5
|
K(lysine) acetyltransferase 5 |
| chr7_+_114562909 | 0.08 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr5_-_159739483 | 0.08 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr11_-_46940074 | 0.08 |
ENST00000378623.1
ENST00000534404.1 |
LRP4
|
low density lipoprotein receptor-related protein 4 |
| chr7_+_143079000 | 0.08 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr15_+_41221536 | 0.08 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr19_-_5978144 | 0.08 |
ENST00000340578.6
ENST00000541471.1 ENST00000591736.1 ENST00000587479.1 |
RANBP3
|
RAN binding protein 3 |
| chr3_-_149375222 | 0.08 |
ENST00000479238.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_+_7452189 | 0.08 |
ENST00000293825.6
|
TNFSF12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chrX_-_128788914 | 0.08 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr19_+_50094866 | 0.08 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr5_+_141016508 | 0.07 |
ENST00000444782.1
ENST00000521367.1 ENST00000297164.3 |
RELL2
|
RELT-like 2 |
| chr5_-_168006324 | 0.07 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr10_+_99258625 | 0.07 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr2_+_220306745 | 0.07 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr14_-_81687575 | 0.07 |
ENST00000434192.2
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr19_-_42724261 | 0.07 |
ENST00000595337.1
|
DEDD2
|
death effector domain containing 2 |
| chr11_-_64578188 | 0.07 |
ENST00000312049.6
ENST00000443283.1 ENST00000315422.4 ENST00000394374.2 |
MEN1
|
multiple endocrine neoplasia I |
| chr17_-_41174424 | 0.07 |
ENST00000355653.3
|
VAT1
|
vesicle amine transport 1 |
| chr19_-_5978090 | 0.07 |
ENST00000592621.1
ENST00000034275.8 ENST00000591092.1 ENST00000591333.1 ENST00000590623.1 ENST00000439268.2 ENST00000587159.1 |
RANBP3
|
RAN binding protein 3 |
| chr19_-_40790729 | 0.07 |
ENST00000423127.1
ENST00000583859.1 ENST00000456441.1 ENST00000452077.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chrX_-_40036520 | 0.07 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr15_-_74726283 | 0.07 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr19_+_15218180 | 0.07 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr20_+_31350184 | 0.07 |
ENST00000328111.2
ENST00000353855.2 ENST00000348286.2 |
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta |
| chr1_+_157963063 | 0.07 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr19_-_5720248 | 0.07 |
ENST00000360614.3
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr6_+_44095347 | 0.07 |
ENST00000323267.6
|
TMEM63B
|
transmembrane protein 63B |
| chr17_+_53342311 | 0.07 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr3_+_49711391 | 0.07 |
ENST00000296456.5
ENST00000449966.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr19_-_55865908 | 0.07 |
ENST00000590900.1
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr17_+_7452336 | 0.07 |
ENST00000293826.4
|
TNFSF12-TNFSF13
|
TNFSF12-TNFSF13 readthrough |
| chr9_-_131038266 | 0.07 |
ENST00000490628.1
ENST00000421699.2 ENST00000450617.1 |
GOLGA2
|
golgin A2 |
| chr12_-_49351148 | 0.07 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr14_+_89029323 | 0.07 |
ENST00000554602.1
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr19_+_8455200 | 0.07 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr3_+_10290596 | 0.07 |
ENST00000448281.2
|
TATDN2
|
TatD DNase domain containing 2 |
| chr20_+_34680698 | 0.07 |
ENST00000447825.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr19_+_13906250 | 0.07 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr15_-_43882353 | 0.07 |
ENST00000453080.1
ENST00000360301.4 ENST00000360135.4 ENST00000417085.1 ENST00000431962.1 ENST00000334933.4 ENST00000381879.4 ENST00000420765.1 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr7_+_100271446 | 0.07 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr22_+_38005033 | 0.07 |
ENST00000447515.1
ENST00000406772.1 ENST00000431745.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr15_+_41952591 | 0.07 |
ENST00000566718.1
ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA
|
MGA, MAX dimerization protein |
| chr1_-_200992827 | 0.07 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr3_+_49711777 | 0.07 |
ENST00000442186.1
ENST00000438011.1 ENST00000457042.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr19_-_40791211 | 0.07 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr19_-_4124079 | 0.07 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr5_-_176037105 | 0.07 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr10_+_103825080 | 0.07 |
ENST00000299238.5
|
HPS6
|
Hermansky-Pudlak syndrome 6 |
| chr19_-_40790993 | 0.07 |
ENST00000596634.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_-_1928621 | 0.07 |
ENST00000331238.6
|
RTN4RL1
|
reticulon 4 receptor-like 1 |
| chr19_+_34287174 | 0.07 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr17_-_74449252 | 0.07 |
ENST00000319380.7
|
UBE2O
|
ubiquitin-conjugating enzyme E2O |
| chr16_-_75299845 | 0.07 |
ENST00000535626.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr1_+_173837488 | 0.07 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr14_+_21945335 | 0.07 |
ENST00000262709.3
ENST00000457430.2 ENST00000448790.2 |
TOX4
|
TOX high mobility group box family member 4 |
| chr19_+_36545781 | 0.06 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr2_-_182521823 | 0.06 |
ENST00000410087.3
ENST00000409440.3 |
CERKL
|
ceramide kinase-like |
| chr17_+_8339340 | 0.06 |
ENST00000580012.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr19_+_8455077 | 0.06 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr7_+_100273736 | 0.06 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr9_+_35605274 | 0.06 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr16_+_75032901 | 0.06 |
ENST00000335325.4
ENST00000320619.6 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chrX_+_48660565 | 0.06 |
ENST00000413163.2
ENST00000441703.1 ENST00000426196.1 |
HDAC6
|
histone deacetylase 6 |
| chr5_+_139781393 | 0.06 |
ENST00000360839.2
ENST00000297183.6 ENST00000421134.1 ENST00000394723.3 ENST00000511151.1 |
ANKHD1
|
ankyrin repeat and KH domain containing 1 |
| chr16_+_75033210 | 0.06 |
ENST00000566250.1
ENST00000567962.1 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr19_-_44259136 | 0.06 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr6_-_31926208 | 0.06 |
ENST00000454913.1
ENST00000436289.2 |
NELFE
|
negative elongation factor complex member E |
| chr11_-_62380199 | 0.06 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr1_-_234745234 | 0.06 |
ENST00000366610.3
ENST00000366609.3 |
IRF2BP2
|
interferon regulatory factor 2 binding protein 2 |
| chr17_-_46703826 | 0.06 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr11_+_45918092 | 0.06 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr20_-_48099182 | 0.06 |
ENST00000371741.4
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr11_+_65837907 | 0.06 |
ENST00000320580.4
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr5_-_159739532 | 0.06 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr1_+_38273818 | 0.06 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr20_-_20693131 | 0.06 |
ENST00000202677.7
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr17_-_7297833 | 0.06 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr6_+_41514305 | 0.06 |
ENST00000409208.1
ENST00000373057.3 |
FOXP4
|
forkhead box P4 |
| chr6_+_24495067 | 0.06 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr1_+_114472481 | 0.06 |
ENST00000369555.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr6_-_31697563 | 0.06 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr5_+_133861339 | 0.06 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr9_-_6645628 | 0.06 |
ENST00000321612.6
|
GLDC
|
glycine dehydrogenase (decarboxylating) |
| chr4_+_8201091 | 0.06 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr19_+_38879061 | 0.06 |
ENST00000587013.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr8_-_145013711 | 0.06 |
ENST00000345136.3
|
PLEC
|
plectin |
| chr11_+_64851729 | 0.06 |
ENST00000526791.1
ENST00000526945.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr19_-_40324255 | 0.06 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.2 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.3 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.2 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0072034 | primary prostatic bud elongation(GO:0060516) bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) renal vesicle induction(GO:0072034) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0072069 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.2 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.3 | GO:1901166 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.2 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:1903911 | positive regulation of receptor clustering(GO:1903911) |
| 0.0 | 0.1 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.1 | GO:0046449 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.3 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.0 | GO:1902336 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.0 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.0 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.0 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.0 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.0 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0060235 | embryonic retina morphogenesis in camera-type eye(GO:0060059) lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.0 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.2 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.0 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.1 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |