Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
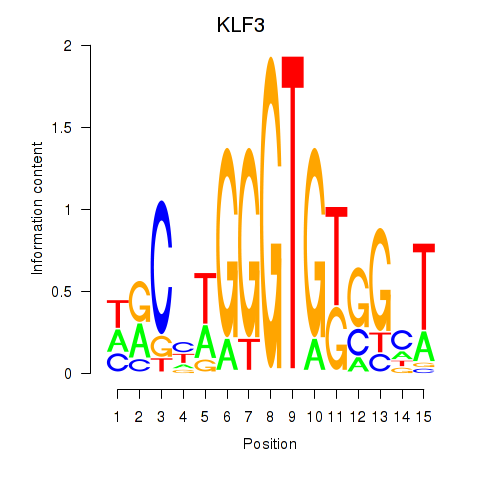
Results for KLF3
Z-value: 0.87
Transcription factors associated with KLF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF3
|
ENSG00000109787.8 | Kruppel like factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF3 | hg19_v2_chr4_+_38665810_38665827 | -0.23 | 6.6e-01 | Click! |
Activity profile of KLF3 motif
Sorted Z-values of KLF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_46492927 | 0.58 |
ENST00000599569.1
|
AP001579.1
|
Uncharacterized protein |
| chr19_+_54135310 | 0.54 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chr20_+_36531544 | 0.49 |
ENST00000448944.1
|
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr3_+_49449636 | 0.45 |
ENST00000273590.3
|
TCTA
|
T-cell leukemia translocation altered |
| chr8_-_123793048 | 0.44 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr5_+_133842243 | 0.42 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr19_-_44008863 | 0.42 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr7_+_33168856 | 0.40 |
ENST00000432983.1
|
BBS9
|
Bardet-Biedl syndrome 9 |
| chr13_-_33002151 | 0.39 |
ENST00000495479.1
ENST00000343281.4 ENST00000464470.1 ENST00000380139.4 ENST00000380133.2 |
N4BP2L1
|
NEDD4 binding protein 2-like 1 |
| chr16_+_8715536 | 0.39 |
ENST00000563958.1
ENST00000381920.3 ENST00000564554.1 |
METTL22
|
methyltransferase like 22 |
| chr6_-_31651817 | 0.38 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr6_+_35744367 | 0.37 |
ENST00000360454.2
ENST00000403376.3 |
CLPSL2
|
colipase-like 2 |
| chr19_-_10464570 | 0.36 |
ENST00000529739.1
|
TYK2
|
tyrosine kinase 2 |
| chr19_+_16308711 | 0.33 |
ENST00000429941.2
ENST00000444449.2 ENST00000589822.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr18_+_46066359 | 0.32 |
ENST00000587752.1
ENST00000588345.1 |
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr17_+_73996987 | 0.31 |
ENST00000588812.1
ENST00000448471.1 |
CDK3
|
cyclin-dependent kinase 3 |
| chr11_+_119038897 | 0.31 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr19_+_36603662 | 0.31 |
ENST00000586670.1
|
OVOL3
|
ovo-like zinc finger 3 |
| chr17_+_79859985 | 0.30 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr7_-_752577 | 0.29 |
ENST00000544935.1
ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr1_+_19967014 | 0.29 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr11_-_65793948 | 0.29 |
ENST00000312106.5
|
CATSPER1
|
cation channel, sperm associated 1 |
| chr1_-_9953295 | 0.29 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr11_-_72432950 | 0.28 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr19_+_16308659 | 0.28 |
ENST00000590263.1
ENST00000590756.1 ENST00000541844.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr17_+_27181956 | 0.28 |
ENST00000254928.5
ENST00000580917.2 |
ERAL1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr19_-_4455290 | 0.28 |
ENST00000394765.3
ENST00000592515.1 |
UBXN6
|
UBX domain protein 6 |
| chr19_+_41860047 | 0.28 |
ENST00000604123.1
|
TMEM91
|
transmembrane protein 91 |
| chr1_+_36789335 | 0.27 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr19_+_39897943 | 0.27 |
ENST00000600033.1
|
ZFP36
|
ZFP36 ring finger protein |
| chr19_+_35630344 | 0.26 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_-_16008880 | 0.26 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr15_+_42565393 | 0.26 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr3_-_50383096 | 0.26 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr19_+_37808778 | 0.25 |
ENST00000544914.1
ENST00000589188.1 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr17_+_6939362 | 0.25 |
ENST00000308027.6
|
SLC16A13
|
solute carrier family 16, member 13 |
| chr11_+_59480899 | 0.24 |
ENST00000300150.7
|
STX3
|
syntaxin 3 |
| chr16_+_50727479 | 0.24 |
ENST00000531674.1
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr19_+_677885 | 0.24 |
ENST00000591552.2
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr1_-_45253377 | 0.24 |
ENST00000372207.3
|
BEST4
|
bestrophin 4 |
| chr3_-_196065374 | 0.24 |
ENST00000454715.1
|
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr8_-_144699628 | 0.24 |
ENST00000529048.1
ENST00000529064.1 |
TSTA3
|
tissue specific transplantation antigen P35B |
| chrX_+_133941218 | 0.24 |
ENST00000370784.4
ENST00000370785.3 |
FAM122C
|
family with sequence similarity 122C |
| chr11_+_126139005 | 0.24 |
ENST00000263578.5
ENST00000442061.2 ENST00000532125.1 |
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr19_-_47616992 | 0.23 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr17_+_73750699 | 0.23 |
ENST00000584939.1
|
ITGB4
|
integrin, beta 4 |
| chr1_+_44679159 | 0.23 |
ENST00000315913.5
ENST00000372289.2 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr9_-_16705069 | 0.23 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr19_+_14492217 | 0.23 |
ENST00000587606.1
ENST00000586517.1 ENST00000591080.1 |
CD97
|
CD97 molecule |
| chr5_-_176433565 | 0.23 |
ENST00000428382.2
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr22_-_36635225 | 0.22 |
ENST00000529194.1
|
APOL2
|
apolipoprotein L, 2 |
| chr15_-_29561979 | 0.22 |
ENST00000332303.4
|
NDNL2
|
necdin-like 2 |
| chr2_+_74881355 | 0.22 |
ENST00000357877.2
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr20_+_55966444 | 0.22 |
ENST00000356208.5
ENST00000440234.2 |
RBM38
|
RNA binding motif protein 38 |
| chr11_+_119039069 | 0.22 |
ENST00000422249.1
|
NLRX1
|
NLR family member X1 |
| chr7_+_870547 | 0.22 |
ENST00000457598.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr17_+_80816395 | 0.22 |
ENST00000576160.2
ENST00000571712.1 |
TBCD
|
tubulin folding cofactor D |
| chr17_+_73089382 | 0.22 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr12_-_57634475 | 0.21 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr7_+_73703728 | 0.21 |
ENST00000361545.5
ENST00000223398.6 |
CLIP2
|
CAP-GLY domain containing linker protein 2 |
| chr11_+_72281681 | 0.21 |
ENST00000450804.3
|
RP11-169D4.1
|
RP11-169D4.1 |
| chr1_+_23695680 | 0.21 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr1_+_44679113 | 0.21 |
ENST00000361745.6
ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr12_+_7013897 | 0.21 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr19_-_14016877 | 0.21 |
ENST00000454313.1
ENST00000591586.1 ENST00000346736.2 |
C19orf57
|
chromosome 19 open reading frame 57 |
| chr19_-_49565254 | 0.21 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr5_-_767034 | 0.21 |
ENST00000382776.4
ENST00000508859.2 |
ZDHHC11B
|
zinc finger, DHHC-type containing 11B |
| chr4_-_190948409 | 0.20 |
ENST00000504750.1
ENST00000378763.1 |
FRG2
|
FSHD region gene 2 |
| chr20_-_1600642 | 0.20 |
ENST00000381603.3
ENST00000381605.4 ENST00000279477.7 ENST00000568365.1 ENST00000564763.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr20_+_62666902 | 0.20 |
ENST00000431158.1
|
LINC00176
|
long intergenic non-protein coding RNA 176 |
| chr16_+_66638003 | 0.20 |
ENST00000562357.1
ENST00000360086.4 ENST00000562707.1 ENST00000361909.4 ENST00000460097.1 ENST00000565666.1 |
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr16_+_2213530 | 0.20 |
ENST00000567645.1
|
TRAF7
|
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
| chr16_-_57837129 | 0.20 |
ENST00000562984.1
ENST00000564891.1 |
KIFC3
|
kinesin family member C3 |
| chr12_+_122326662 | 0.20 |
ENST00000261817.2
ENST00000538613.1 ENST00000542602.1 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr22_-_36850991 | 0.20 |
ENST00000442579.1
|
RP5-1119A7.14
|
RP5-1119A7.14 |
| chr10_-_51486327 | 0.20 |
ENST00000374095.5
|
AGAP7
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 7 |
| chr12_-_49351148 | 0.20 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr3_-_196065248 | 0.19 |
ENST00000446879.1
ENST00000273695.3 |
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr7_+_872107 | 0.19 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr11_+_6226782 | 0.19 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr1_-_24126051 | 0.19 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr12_+_122326630 | 0.19 |
ENST00000541212.1
ENST00000340175.5 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr15_-_78933567 | 0.18 |
ENST00000261751.3
ENST00000412074.2 |
CHRNB4
|
cholinergic receptor, nicotinic, beta 4 (neuronal) |
| chr22_+_29702996 | 0.18 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr15_-_28344439 | 0.18 |
ENST00000431101.1
ENST00000445578.1 ENST00000353809.5 ENST00000382996.2 ENST00000354638.3 |
OCA2
|
oculocutaneous albinism II |
| chr16_-_86588627 | 0.18 |
ENST00000565482.1
ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing |
| chr2_+_217498105 | 0.18 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr2_+_74881398 | 0.18 |
ENST00000339773.5
ENST00000434486.1 |
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr17_+_7452336 | 0.18 |
ENST00000293826.4
|
TNFSF12-TNFSF13
|
TNFSF12-TNFSF13 readthrough |
| chr11_+_126173647 | 0.18 |
ENST00000263579.4
|
DCPS
|
decapping enzyme, scavenger |
| chr7_-_73153178 | 0.18 |
ENST00000437775.2
ENST00000222800.3 |
ABHD11
|
abhydrolase domain containing 11 |
| chr4_+_2794750 | 0.18 |
ENST00000452765.2
ENST00000389838.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr16_+_69333585 | 0.18 |
ENST00000570054.2
|
RP11-343C2.11
|
Uncharacterized protein |
| chr19_-_14064114 | 0.18 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr2_+_177502438 | 0.18 |
ENST00000443670.1
|
AC017048.4
|
long intergenic non-protein coding RNA 1117 |
| chr17_-_76870126 | 0.18 |
ENST00000586057.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr17_-_62207485 | 0.18 |
ENST00000433197.3
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr7_+_36192758 | 0.18 |
ENST00000242108.4
|
EEPD1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr16_-_67514982 | 0.18 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr1_+_17944832 | 0.17 |
ENST00000167825.4
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr17_+_79934670 | 0.17 |
ENST00000581484.1
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr9_-_140115775 | 0.17 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr20_+_61299155 | 0.17 |
ENST00000451793.1
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1 |
| chr22_+_40573921 | 0.17 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr2_-_74710078 | 0.17 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chr19_+_14492247 | 0.17 |
ENST00000357355.3
ENST00000592261.2 ENST00000242786.5 |
CD97
|
CD97 molecule |
| chr16_+_84209738 | 0.17 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr16_-_57832004 | 0.17 |
ENST00000562503.1
|
KIFC3
|
kinesin family member C3 |
| chr11_+_844406 | 0.17 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr15_+_67430339 | 0.17 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr19_-_4722705 | 0.17 |
ENST00000598360.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chrX_-_152245978 | 0.17 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr12_-_54121261 | 0.17 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr15_+_75074410 | 0.17 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr2_+_128458514 | 0.17 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr22_+_19706958 | 0.17 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr8_+_123793633 | 0.16 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr22_-_51017084 | 0.16 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr19_-_4723761 | 0.16 |
ENST00000597849.1
ENST00000598800.1 ENST00000602161.1 ENST00000597726.1 ENST00000601130.1 ENST00000262960.9 |
DPP9
|
dipeptidyl-peptidase 9 |
| chr14_-_21566731 | 0.16 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr16_-_57513657 | 0.16 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr11_+_65383227 | 0.16 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr2_-_24307635 | 0.16 |
ENST00000313482.4
|
TP53I3
|
tumor protein p53 inducible protein 3 |
| chr7_-_99717463 | 0.16 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr12_+_48516357 | 0.16 |
ENST00000549022.1
ENST00000547587.1 ENST00000312352.7 |
PFKM
|
phosphofructokinase, muscle |
| chr19_-_2740036 | 0.16 |
ENST00000586572.1
ENST00000269740.4 |
AC006538.4
SLC39A3
|
Uncharacterized protein solute carrier family 39 (zinc transporter), member 3 |
| chr10_-_118429461 | 0.16 |
ENST00000588184.1
ENST00000369210.3 |
C10orf82
|
chromosome 10 open reading frame 82 |
| chr20_-_52687030 | 0.16 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chrX_+_70364667 | 0.16 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr10_+_82009466 | 0.16 |
ENST00000356374.4
|
AL359195.1
|
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
| chr2_+_71295766 | 0.16 |
ENST00000533981.1
|
NAGK
|
N-acetylglucosamine kinase |
| chr17_+_2699697 | 0.16 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr7_+_36192855 | 0.16 |
ENST00000534978.1
|
EEPD1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr11_-_65430251 | 0.16 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr16_+_84209539 | 0.16 |
ENST00000569735.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chrX_+_152338301 | 0.15 |
ENST00000453825.2
|
PNMA6A
|
paraneoplastic Ma antigen family member 6A |
| chr11_+_3875757 | 0.15 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr19_-_40324767 | 0.15 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr2_-_128408105 | 0.15 |
ENST00000409254.1
|
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr3_+_186648507 | 0.15 |
ENST00000458216.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_-_19302931 | 0.15 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr3_+_187957646 | 0.15 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_107582775 | 0.15 |
ENST00000305991.2
|
SLN
|
sarcolipin |
| chr17_-_43138463 | 0.15 |
ENST00000310604.4
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr15_+_75074915 | 0.15 |
ENST00000567123.1
ENST00000569462.1 |
CSK
|
c-src tyrosine kinase |
| chr22_-_36635563 | 0.15 |
ENST00000451256.2
|
APOL2
|
apolipoprotein L, 2 |
| chr3_+_9773409 | 0.15 |
ENST00000433861.2
ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr1_-_1655713 | 0.15 |
ENST00000401096.2
ENST00000404249.3 ENST00000357760.2 ENST00000358779.5 ENST00000378633.1 ENST00000378635.3 |
CDK11A
|
cyclin-dependent kinase 11A |
| chr1_-_206671061 | 0.15 |
ENST00000367119.1
|
C1orf147
|
chromosome 1 open reading frame 147 |
| chr22_+_35776828 | 0.15 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr2_+_71295717 | 0.15 |
ENST00000418807.3
ENST00000443872.2 |
NAGK
|
N-acetylglucosamine kinase |
| chr19_+_47616682 | 0.15 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr15_+_75074385 | 0.15 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr1_+_2004901 | 0.15 |
ENST00000400921.2
|
PRKCZ
|
protein kinase C, zeta |
| chr4_-_90757364 | 0.15 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_65647280 | 0.14 |
ENST00000307886.3
ENST00000528419.1 ENST00000526034.1 |
CTSW
|
cathepsin W |
| chr10_+_51748404 | 0.14 |
ENST00000412531.3
|
AGAP6
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 6 |
| chr19_+_17337027 | 0.14 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr4_+_189376725 | 0.14 |
ENST00000510832.1
ENST00000510005.1 ENST00000503458.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr14_+_105147464 | 0.14 |
ENST00000540171.2
|
RP11-982M15.6
|
RP11-982M15.6 |
| chr2_+_220143989 | 0.14 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr10_-_134145321 | 0.14 |
ENST00000368625.4
ENST00000368619.3 ENST00000456004.1 ENST00000368620.2 |
STK32C
|
serine/threonine kinase 32C |
| chr11_-_65626797 | 0.14 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr19_+_39279838 | 0.14 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr2_+_71295733 | 0.14 |
ENST00000443938.2
ENST00000244204.6 |
NAGK
|
N-acetylglucosamine kinase |
| chr17_-_76732928 | 0.14 |
ENST00000589768.1
|
CYTH1
|
cytohesin 1 |
| chr1_+_117544366 | 0.14 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr19_-_33182616 | 0.14 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr7_+_100466433 | 0.14 |
ENST00000429658.1
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr16_+_67233412 | 0.14 |
ENST00000477898.1
|
ELMO3
|
engulfment and cell motility 3 |
| chr8_-_13134045 | 0.14 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr12_-_54121212 | 0.14 |
ENST00000548263.1
ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr10_+_71211212 | 0.14 |
ENST00000373290.2
|
TSPAN15
|
tetraspanin 15 |
| chr16_-_30381580 | 0.14 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr11_+_68816356 | 0.14 |
ENST00000294309.3
ENST00000542467.1 |
TPCN2
|
two pore segment channel 2 |
| chr17_-_39623681 | 0.14 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr7_-_73153161 | 0.14 |
ENST00000395147.4
|
ABHD11
|
abhydrolase domain containing 11 |
| chr17_-_8198636 | 0.14 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr16_-_57831914 | 0.14 |
ENST00000421376.2
|
KIFC3
|
kinesin family member C3 |
| chr1_+_17914907 | 0.14 |
ENST00000375420.3
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr11_-_65626753 | 0.14 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr16_-_79804435 | 0.14 |
ENST00000562921.1
ENST00000566729.1 |
RP11-345M22.2
|
RP11-345M22.2 |
| chr19_-_2739992 | 0.14 |
ENST00000545664.1
ENST00000589363.1 ENST00000455372.2 |
SLC39A3
|
solute carrier family 39 (zinc transporter), member 3 |
| chr12_+_116997186 | 0.13 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr16_+_31483374 | 0.13 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr17_-_48278983 | 0.13 |
ENST00000225964.5
|
COL1A1
|
collagen, type I, alpha 1 |
| chrX_-_153599578 | 0.13 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr19_+_14017116 | 0.13 |
ENST00000589606.1
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr7_+_73245193 | 0.13 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr16_+_29832634 | 0.13 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr9_+_131902283 | 0.13 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_+_45477819 | 0.13 |
ENST00000246337.4
|
UROD
|
uroporphyrinogen decarboxylase |
| chr17_+_37030127 | 0.13 |
ENST00000419929.1
|
LASP1
|
LIM and SH3 protein 1 |
| chr9_+_140149625 | 0.13 |
ENST00000343053.4
|
NELFB
|
negative elongation factor complex member B |
| chr16_-_57831676 | 0.13 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr7_+_44788430 | 0.13 |
ENST00000457123.1
ENST00000309315.4 |
ZMIZ2
|
zinc finger, MIZ-type containing 2 |
| chr19_+_46850320 | 0.13 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr22_+_21737663 | 0.13 |
ENST00000434111.1
|
RIMBP3B
|
RIMS binding protein 3B |
| chr20_+_9049742 | 0.13 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr17_+_18128896 | 0.13 |
ENST00000316843.4
|
LLGL1
|
lethal giant larvae homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.1 | 0.3 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.2 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 0.4 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.5 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.5 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.3 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.2 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 0.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.3 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.0 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.2 | GO:0043474 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.3 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.3 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.3 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.4 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.2 | GO:1903974 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 | 0.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.3 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.5 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.6 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.0 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.1 | GO:0090034 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0042306 | regulation of protein import into nucleus(GO:0042306) regulation of protein import(GO:1904589) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.0 | GO:0090071 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.0 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.0 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.3 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.5 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.3 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 0.2 | GO:0050577 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.1 | 0.3 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 0.2 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0008124 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.4 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.0 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |