Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
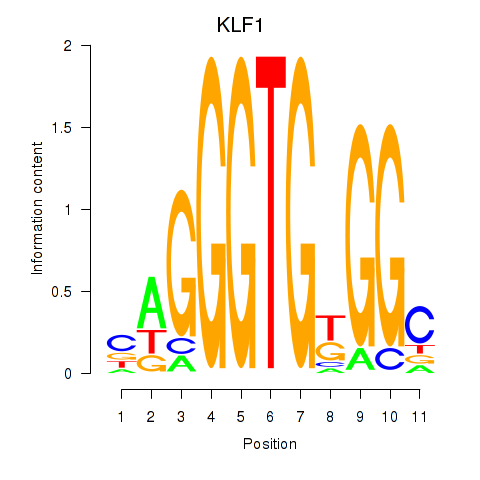
Results for KLF1
Z-value: 1.29
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.4 | Kruppel like factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF1 | hg19_v2_chr19_-_12997995_12998021 | 0.48 | 3.4e-01 | Click! |
Activity profile of KLF1 motif
Sorted Z-values of KLF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_58145889 | 1.48 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr2_+_74212073 | 1.45 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr3_+_49449636 | 0.96 |
ENST00000273590.3
|
TCTA
|
T-cell leukemia translocation altered |
| chrX_-_16888276 | 0.85 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr17_+_7210921 | 0.80 |
ENST00000573542.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr19_-_44008863 | 0.74 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr1_-_145589424 | 0.72 |
ENST00000334513.5
|
NUDT17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr10_+_17272608 | 0.71 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr17_+_7210852 | 0.70 |
ENST00000576930.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_-_58212487 | 0.68 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr14_+_21569245 | 0.68 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr3_-_50383096 | 0.67 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr17_+_46131912 | 0.65 |
ENST00000584634.1
ENST00000580050.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr15_-_44069513 | 0.63 |
ENST00000433927.1
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr20_-_62168672 | 0.57 |
ENST00000217185.2
|
PTK6
|
protein tyrosine kinase 6 |
| chrX_+_47077680 | 0.57 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr8_-_123793048 | 0.57 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr11_+_62475130 | 0.56 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr2_+_206950095 | 0.55 |
ENST00000435627.1
|
AC007383.3
|
AC007383.3 |
| chr14_-_100841794 | 0.54 |
ENST00000556295.1
ENST00000554820.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr2_-_26205340 | 0.54 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr17_-_7493390 | 0.52 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr13_-_74708372 | 0.52 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr19_+_54960358 | 0.51 |
ENST00000439657.1
ENST00000376514.2 ENST00000376526.4 ENST00000436479.1 |
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr17_+_7210898 | 0.51 |
ENST00000572815.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr8_-_145018080 | 0.51 |
ENST00000354589.3
|
PLEC
|
plectin |
| chr6_-_38607673 | 0.50 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chrX_+_47077632 | 0.50 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr20_-_52687030 | 0.50 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr15_+_76352178 | 0.50 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr6_-_38607628 | 0.49 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr17_-_46716647 | 0.48 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr11_-_1606513 | 0.47 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr22_-_20138302 | 0.47 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr2_+_75185619 | 0.45 |
ENST00000483063.1
|
POLE4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr14_-_100841670 | 0.45 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr19_+_54960790 | 0.45 |
ENST00000443957.1
|
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr5_+_133562095 | 0.44 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr17_+_7210294 | 0.44 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_+_57943781 | 0.43 |
ENST00000455537.2
ENST00000286452.5 |
KIF5A
|
kinesin family member 5A |
| chr2_-_26205550 | 0.43 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
| chr14_+_77425972 | 0.43 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr20_+_825275 | 0.42 |
ENST00000541082.1
|
FAM110A
|
family with sequence similarity 110, member A |
| chr16_-_30597000 | 0.42 |
ENST00000470110.1
ENST00000395216.2 |
ZNF785
|
zinc finger protein 785 |
| chr9_-_33264676 | 0.42 |
ENST00000472232.3
ENST00000379704.2 |
BAG1
|
BCL2-associated athanogene |
| chr17_-_1619535 | 0.41 |
ENST00000573075.1
ENST00000574306.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr1_+_156096336 | 0.41 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr9_-_33264557 | 0.41 |
ENST00000473781.1
ENST00000488499.1 |
BAG1
|
BCL2-associated athanogene |
| chr17_+_1633755 | 0.41 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chr16_+_50313426 | 0.40 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr11_+_63655987 | 0.39 |
ENST00000509502.2
ENST00000512060.1 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr12_-_100378392 | 0.39 |
ENST00000549866.1
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr14_+_24783906 | 0.38 |
ENST00000396782.2
|
LTB4R
|
leukotriene B4 receptor |
| chr17_-_34122596 | 0.37 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr1_+_167190066 | 0.37 |
ENST00000367866.2
ENST00000429375.2 ENST00000452019.1 ENST00000420254.3 ENST00000541643.3 |
POU2F1
|
POU class 2 homeobox 1 |
| chr16_+_31483451 | 0.36 |
ENST00000565360.1
ENST00000361773.3 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr15_-_78112553 | 0.36 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr12_-_58131931 | 0.36 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr11_-_65314905 | 0.35 |
ENST00000527339.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr3_-_105588231 | 0.35 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr11_+_47608198 | 0.35 |
ENST00000356737.2
ENST00000538490.1 |
FAM180B
|
family with sequence similarity 180, member B |
| chr9_+_74764278 | 0.34 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr8_+_21912328 | 0.34 |
ENST00000432128.1
ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN
|
dematin actin binding protein |
| chr19_+_38880252 | 0.34 |
ENST00000586301.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr16_+_31483374 | 0.34 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr9_-_130616915 | 0.33 |
ENST00000344849.3
|
ENG
|
endoglin |
| chr4_-_90756769 | 0.33 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_+_36119975 | 0.33 |
ENST00000589559.1
ENST00000360475.4 |
RBM42
|
RNA binding motif protein 42 |
| chrX_-_54824673 | 0.33 |
ENST00000218436.6
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr15_+_75074410 | 0.33 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr16_+_30078811 | 0.33 |
ENST00000564688.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr14_+_77228532 | 0.33 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr15_+_75074385 | 0.33 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr19_+_36120009 | 0.33 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr12_+_116997186 | 0.33 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr2_-_197458271 | 0.33 |
ENST00000427457.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_-_86564696 | 0.32 |
ENST00000437769.1
|
REEP1
|
receptor accessory protein 1 |
| chr5_+_133859996 | 0.32 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr8_+_123793633 | 0.32 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr19_+_41698927 | 0.32 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr17_-_27188984 | 0.32 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr11_-_119252425 | 0.32 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr16_-_4466622 | 0.31 |
ENST00000570645.1
ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7
|
coronin 7 |
| chr17_-_79520135 | 0.31 |
ENST00000541246.1
ENST00000544302.1 |
C17orf70
|
chromosome 17 open reading frame 70 |
| chrX_+_140677562 | 0.31 |
ENST00000370518.3
|
SPANXA2
|
SPANX family, member A2 |
| chr10_-_47173994 | 0.31 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr19_+_35607166 | 0.31 |
ENST00000604255.1
ENST00000346446.5 ENST00000344013.6 ENST00000603449.1 ENST00000406988.1 ENST00000605550.1 ENST00000604804.1 ENST00000605552.1 |
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr1_-_24126051 | 0.31 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr6_+_31691121 | 0.31 |
ENST00000480039.1
ENST00000375810.4 ENST00000375805.2 ENST00000375809.3 ENST00000375804.2 ENST00000375814.3 ENST00000375806.2 |
C6orf25
|
chromosome 6 open reading frame 25 |
| chr1_+_43148059 | 0.30 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr12_-_56693758 | 0.30 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr1_+_11866270 | 0.30 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr19_+_42788172 | 0.30 |
ENST00000160740.3
|
CIC
|
capicua transcriptional repressor |
| chr19_-_39340563 | 0.30 |
ENST00000601813.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr3_-_71114066 | 0.30 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr19_+_39574945 | 0.30 |
ENST00000331256.5
|
PAPL
|
Iron/zinc purple acid phosphatase-like protein |
| chr9_-_130617029 | 0.30 |
ENST00000373203.4
|
ENG
|
endoglin |
| chr6_-_31763276 | 0.30 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr18_+_29027696 | 0.30 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr17_+_46125685 | 0.30 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr11_-_64014379 | 0.29 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr22_+_38071615 | 0.29 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr6_+_33359582 | 0.29 |
ENST00000450504.1
|
KIFC1
|
kinesin family member C1 |
| chr7_-_35840198 | 0.29 |
ENST00000412856.1
ENST00000437235.3 ENST00000424194.1 |
AC007551.3
|
AC007551.3 |
| chr12_-_57443886 | 0.29 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr15_-_72521017 | 0.29 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr5_-_89770171 | 0.29 |
ENST00000514906.1
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr1_+_17914907 | 0.29 |
ENST00000375420.3
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr2_+_219524473 | 0.28 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr17_+_39845134 | 0.28 |
ENST00000591776.1
ENST00000469257.1 |
EIF1
|
eukaryotic translation initiation factor 1 |
| chr17_-_27405875 | 0.28 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr11_+_65339820 | 0.28 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.1 |
FAM89B
|
family with sequence similarity 89, member B |
| chr17_-_79519403 | 0.28 |
ENST00000327787.8
ENST00000537152.1 |
C17orf70
|
chromosome 17 open reading frame 70 |
| chr1_+_33231221 | 0.28 |
ENST00000294521.3
|
KIAA1522
|
KIAA1522 |
| chr17_-_26220366 | 0.28 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr19_-_47616992 | 0.28 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr11_+_65647280 | 0.28 |
ENST00000307886.3
ENST00000528419.1 ENST00000526034.1 |
CTSW
|
cathepsin W |
| chr19_+_1077393 | 0.28 |
ENST00000590577.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr1_+_11866207 | 0.27 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr5_+_321810 | 0.27 |
ENST00000514523.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr19_-_10676666 | 0.27 |
ENST00000539027.1
ENST00000543682.1 ENST00000361821.5 ENST00000312962.6 |
KRI1
|
KRI1 homolog (S. cerevisiae) |
| chr12_-_63544718 | 0.27 |
ENST00000299178.2
|
AVPR1A
|
arginine vasopressin receptor 1A |
| chr6_-_30654977 | 0.27 |
ENST00000399199.3
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_-_39395165 | 0.27 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr17_-_74023291 | 0.27 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr17_-_74023474 | 0.27 |
ENST00000301607.3
|
EVPL
|
envoplakin |
| chr4_+_39640787 | 0.27 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr15_+_88795953 | 0.26 |
ENST00000569588.1
|
NTRK3-AS1
|
NTRK3 antisense RNA 1 |
| chr13_+_24553933 | 0.26 |
ENST00000424834.2
ENST00000439928.2 |
SPATA13
RP11-309I15.1
|
spermatogenesis associated 13 RP11-309I15.1 |
| chr5_+_71403061 | 0.26 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr19_+_42806250 | 0.26 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr19_+_18668572 | 0.26 |
ENST00000540691.1
ENST00000539106.1 ENST00000222307.4 |
KXD1
|
KxDL motif containing 1 |
| chr1_+_27189631 | 0.26 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr16_+_67233007 | 0.26 |
ENST00000360833.1
ENST00000393997.2 |
ELMO3
|
engulfment and cell motility 3 |
| chr4_-_926161 | 0.26 |
ENST00000511163.1
|
GAK
|
cyclin G associated kinase |
| chr11_+_1889880 | 0.26 |
ENST00000405957.2
|
LSP1
|
lymphocyte-specific protein 1 |
| chr17_-_39538550 | 0.25 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr19_+_42363917 | 0.25 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr6_+_106546808 | 0.25 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr17_+_37030127 | 0.25 |
ENST00000419929.1
|
LASP1
|
LIM and SH3 protein 1 |
| chr17_-_2117600 | 0.25 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chrX_+_47078069 | 0.25 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr1_-_24126023 | 0.25 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr8_+_145490549 | 0.25 |
ENST00000340695.2
|
SCXA
|
scleraxis homolog A (mouse) |
| chr1_-_94147385 | 0.24 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr19_+_41699135 | 0.24 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr1_-_159893507 | 0.24 |
ENST00000368096.1
|
TAGLN2
|
transgelin 2 |
| chr9_-_35732362 | 0.24 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chr17_-_1389228 | 0.24 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr6_-_131291572 | 0.24 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr19_+_18451439 | 0.24 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr1_-_161039753 | 0.23 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr17_-_41174367 | 0.23 |
ENST00000587173.1
|
VAT1
|
vesicle amine transport 1 |
| chr22_+_20119320 | 0.23 |
ENST00000334554.7
ENST00000320602.7 ENST00000405930.3 |
ZDHHC8
|
zinc finger, DHHC-type containing 8 |
| chr3_-_45838011 | 0.23 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr10_-_94003003 | 0.23 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr11_+_65657875 | 0.23 |
ENST00000312579.2
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr14_+_32414059 | 0.23 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr14_-_21566731 | 0.22 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr11_+_71903169 | 0.22 |
ENST00000393676.3
|
FOLR1
|
folate receptor 1 (adult) |
| chr9_-_135819987 | 0.22 |
ENST00000298552.3
ENST00000403810.1 |
TSC1
|
tuberous sclerosis 1 |
| chr17_-_56065540 | 0.22 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr1_-_98510843 | 0.22 |
ENST00000413670.2
ENST00000538428.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr19_-_36523529 | 0.21 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr2_-_70944855 | 0.21 |
ENST00000415348.1
|
ADD2
|
adducin 2 (beta) |
| chr22_-_36357671 | 0.21 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr17_-_46703826 | 0.21 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr9_-_140353748 | 0.21 |
ENST00000371472.2
ENST00000371475.3 ENST00000265663.7 ENST00000437259.1 ENST00000392812.4 ENST00000371474.3 ENST00000371473.3 |
NSMF
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr19_-_42916499 | 0.21 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr17_-_40575535 | 0.21 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr7_+_127228399 | 0.21 |
ENST00000000233.5
ENST00000415666.1 |
ARF5
|
ADP-ribosylation factor 5 |
| chr5_+_140514782 | 0.21 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr2_-_219524193 | 0.21 |
ENST00000450560.1
ENST00000449707.1 ENST00000432460.1 ENST00000411696.2 |
ZNF142
|
zinc finger protein 142 |
| chr2_+_242127924 | 0.21 |
ENST00000402530.3
ENST00000274979.8 ENST00000402430.3 |
ANO7
|
anoctamin 7 |
| chr17_+_46131843 | 0.21 |
ENST00000577411.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr8_+_22423168 | 0.21 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_+_153747746 | 0.21 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr1_-_161039647 | 0.21 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr1_-_117664317 | 0.21 |
ENST00000256649.4
ENST00000369464.3 ENST00000485032.1 |
TRIM45
|
tripartite motif containing 45 |
| chr12_+_26274917 | 0.21 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr8_+_21911054 | 0.21 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr17_+_40996590 | 0.21 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr17_-_1619568 | 0.20 |
ENST00000571595.1
|
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr6_+_42952237 | 0.20 |
ENST00000485511.1
ENST00000394110.3 ENST00000472118.1 ENST00000461010.1 |
PPP2R5D
|
protein phosphatase 2, regulatory subunit B', delta |
| chr1_-_11866034 | 0.20 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr20_+_35974532 | 0.20 |
ENST00000373578.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr16_+_30075783 | 0.20 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_-_45837959 | 0.20 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr3_-_105587879 | 0.20 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chrX_-_48814810 | 0.20 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr15_-_89878025 | 0.20 |
ENST00000268124.5
ENST00000442287.2 |
POLG
|
polymerase (DNA directed), gamma |
| chr5_-_115910630 | 0.20 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_+_140480083 | 0.20 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr19_-_55791540 | 0.20 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr19_-_55791058 | 0.20 |
ENST00000587959.1
ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr17_-_7991021 | 0.19 |
ENST00000319144.4
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type |
| chrX_+_70503526 | 0.19 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr6_+_43027595 | 0.19 |
ENST00000259708.3
ENST00000472792.1 ENST00000479388.1 ENST00000460283.1 ENST00000394056.2 |
KLC4
|
kinesin light chain 4 |
| chr11_-_119252359 | 0.19 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr12_+_57998400 | 0.19 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.6 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.2 | 1.8 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.2 | 0.5 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 1.0 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.0 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.4 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.4 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.8 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.6 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.8 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.8 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.4 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.4 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.2 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.3 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of cellular pH reduction(GO:0032849) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.3 | GO:0009093 | cysteine biosynthetic process from serine(GO:0006535) cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.3 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.5 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.1 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 0.3 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.2 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.3 | GO:2000771 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0090650 | rRNA transport(GO:0051029) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.4 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.6 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 1.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.3 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0044115 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.2 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 1.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:0003096 | renal sodium ion transport(GO:0003096) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.8 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.3 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 1.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0093001 | glycolytic process through glucose-1-phosphate(GO:0061622) glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.3 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 1.0 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.3 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 0.6 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.2 | 2.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.4 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.5 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.6 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 2.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.3 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.4 | GO:0004974 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:0050421 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.1 | 0.3 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.2 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.2 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 2.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle binding(GO:0034189) very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.6 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 2.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 2.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |