Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
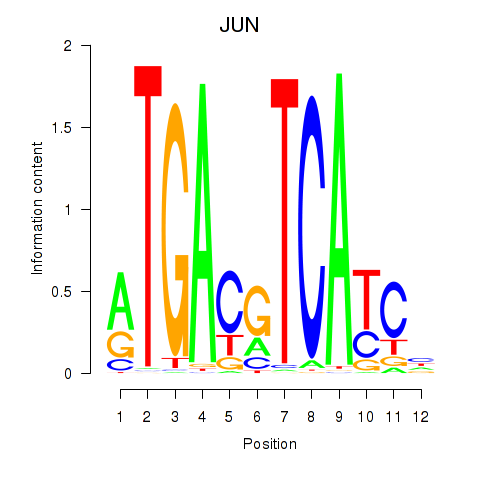
Results for JUN
Z-value: 1.04
Transcription factors associated with JUN
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUN
|
ENSG00000177606.5 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUN | hg19_v2_chr1_-_59249732_59249785 | 0.55 | 2.5e-01 | Click! |
Activity profile of JUN motif
Sorted Z-values of JUN motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_139944396 | 0.98 |
ENST00000514199.1
|
SLC35A4
|
solute carrier family 35, member A4 |
| chr14_+_100485712 | 0.93 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr19_-_50979981 | 0.85 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr5_+_172332220 | 0.81 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr12_+_10365082 | 0.80 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr5_-_175964366 | 0.80 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr19_-_47616992 | 0.76 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr19_+_50979753 | 0.74 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr16_+_2564254 | 0.69 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr11_-_1593150 | 0.68 |
ENST00000397374.3
|
DUSP8
|
dual specificity phosphatase 8 |
| chr5_-_175965008 | 0.68 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chr14_+_93389425 | 0.66 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chrX_+_47342970 | 0.62 |
ENST00000357412.1
|
CXorf24
|
chromosome X open reading frame 24 |
| chr11_-_9482010 | 0.61 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr5_+_139944024 | 0.59 |
ENST00000323146.3
|
SLC35A4
|
solute carrier family 35, member A4 |
| chr14_-_91294472 | 0.59 |
ENST00000555975.1
|
CTD-3035D6.2
|
CTD-3035D6.2 |
| chr5_+_139055021 | 0.58 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr6_-_121655850 | 0.58 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr19_-_46088068 | 0.56 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr19_-_48613987 | 0.56 |
ENST00000596138.1
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr19_-_48614063 | 0.56 |
ENST00000599921.1
ENST00000599111.1 |
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr5_+_139055055 | 0.56 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr17_-_43210580 | 0.56 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr1_-_153917700 | 0.55 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr19_+_19030497 | 0.55 |
ENST00000438170.2
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr19_+_19030478 | 0.55 |
ENST00000247003.4
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr5_-_99870932 | 0.53 |
ENST00000504833.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr19_+_36024310 | 0.52 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr11_+_117070037 | 0.52 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr3_-_12587055 | 0.51 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr6_-_34664612 | 0.51 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr7_+_30174574 | 0.50 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chrX_+_48398053 | 0.50 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr19_+_42772659 | 0.49 |
ENST00000572681.2
|
CIC
|
capicua transcriptional repressor |
| chr11_-_65793948 | 0.48 |
ENST00000312106.5
|
CATSPER1
|
cation channel, sperm associated 1 |
| chr12_-_123380610 | 0.48 |
ENST00000535765.1
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr22_-_19132154 | 0.48 |
ENST00000252137.6
|
DGCR14
|
DiGeorge syndrome critical region gene 14 |
| chr19_+_13056663 | 0.47 |
ENST00000541222.1
ENST00000316856.3 ENST00000586534.1 ENST00000592268.1 |
RAD23A
|
RAD23 homolog A (S. cerevisiae) |
| chr6_-_41888843 | 0.47 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr19_+_7981030 | 0.45 |
ENST00000565886.1
|
TGFBR3L
|
transforming growth factor, beta receptor III-like |
| chr7_-_129592471 | 0.44 |
ENST00000473814.2
ENST00000490974.1 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr3_-_183979251 | 0.44 |
ENST00000296238.3
|
CAMK2N2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr7_-_129592700 | 0.44 |
ENST00000472396.1
ENST00000355621.3 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr17_+_7835419 | 0.43 |
ENST00000576538.1
ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr11_+_12696102 | 0.42 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr5_-_86534822 | 0.42 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr8_-_95274536 | 0.42 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr5_+_133562095 | 0.41 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr3_+_128598433 | 0.41 |
ENST00000308982.7
ENST00000514336.1 |
ACAD9
|
acyl-CoA dehydrogenase family, member 9 |
| chr17_+_7155343 | 0.40 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr21_-_44751903 | 0.40 |
ENST00000450205.1
|
LINC00322
|
long intergenic non-protein coding RNA 322 |
| chr19_-_51014460 | 0.40 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr19_+_47616682 | 0.39 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chrX_-_101771645 | 0.39 |
ENST00000289373.4
|
TMSB15A
|
thymosin beta 15a |
| chr7_+_100551239 | 0.39 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr8_-_21966893 | 0.38 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr3_+_42190714 | 0.38 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr17_+_7155556 | 0.38 |
ENST00000570500.1
ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr2_-_98280383 | 0.38 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr16_+_3068393 | 0.37 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr12_-_6982442 | 0.37 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr1_-_32860020 | 0.37 |
ENST00000527163.1
ENST00000341071.7 ENST00000530485.1 ENST00000446293.2 ENST00000413080.1 ENST00000449308.1 ENST00000526031.1 ENST00000419121.2 ENST00000455895.2 |
BSDC1
|
BSD domain containing 1 |
| chr11_-_8832182 | 0.37 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr16_-_86588627 | 0.37 |
ENST00000565482.1
ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing |
| chr1_+_36024107 | 0.37 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr6_-_31940065 | 0.37 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr19_-_48614033 | 0.36 |
ENST00000354276.3
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr12_-_371994 | 0.36 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr5_-_151066514 | 0.36 |
ENST00000538026.1
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr7_-_45026159 | 0.36 |
ENST00000584327.1
ENST00000438705.3 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr2_+_47043832 | 0.36 |
ENST00000422294.1
|
AC016722.2
|
long intergenic non-protein coding RNA 1119 |
| chr19_-_4717835 | 0.36 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr7_-_20826504 | 0.36 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr11_-_64647144 | 0.35 |
ENST00000359393.2
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH-domain containing 1 |
| chr19_+_49535169 | 0.35 |
ENST00000474913.1
ENST00000359342.6 |
CGB2
|
chorionic gonadotropin, beta polypeptide 2 |
| chr13_+_111972980 | 0.34 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chrX_-_152245978 | 0.34 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr19_-_55574538 | 0.34 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr11_+_122709200 | 0.34 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr2_+_11864458 | 0.34 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr11_+_6226782 | 0.34 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr12_-_52761262 | 0.34 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr19_-_51014345 | 0.34 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr19_-_44100275 | 0.33 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr2_-_74781061 | 0.33 |
ENST00000264094.3
ENST00000393937.2 ENST00000409986.1 |
LOXL3
|
lysyl oxidase-like 3 |
| chrX_+_152240819 | 0.33 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr1_+_28099700 | 0.33 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr7_+_134832808 | 0.33 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr13_+_111855399 | 0.32 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr22_+_22988816 | 0.32 |
ENST00000480559.1
ENST00000448514.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chr9_+_105757590 | 0.32 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr17_-_43209862 | 0.32 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr20_+_3190006 | 0.32 |
ENST00000380113.3
ENST00000455664.2 ENST00000399838.3 |
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr1_-_32264250 | 0.31 |
ENST00000528579.1
|
SPOCD1
|
SPOC domain containing 1 |
| chr4_-_798689 | 0.31 |
ENST00000504062.1
|
CPLX1
|
complexin 1 |
| chr7_+_30174668 | 0.31 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr19_+_36119975 | 0.31 |
ENST00000589559.1
ENST00000360475.4 |
RBM42
|
RNA binding motif protein 42 |
| chr11_+_94439591 | 0.31 |
ENST00000299004.9
|
AMOTL1
|
angiomotin like 1 |
| chr7_-_45026200 | 0.31 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr12_+_52668394 | 0.31 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chrX_+_153046456 | 0.31 |
ENST00000393786.3
ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3
|
SRSF protein kinase 3 |
| chr6_-_3157760 | 0.31 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr12_+_10365404 | 0.30 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr17_+_33448593 | 0.30 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr16_+_30662085 | 0.30 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr14_-_24732738 | 0.30 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr11_-_119187826 | 0.30 |
ENST00000264036.4
|
MCAM
|
melanoma cell adhesion molecule |
| chr10_+_99496872 | 0.30 |
ENST00000337540.7
ENST00000357540.4 ENST00000370613.3 ENST00000370610.3 ENST00000393677.4 ENST00000453958.2 ENST00000359980.3 |
ZFYVE27
|
zinc finger, FYVE domain containing 27 |
| chr9_-_140115775 | 0.30 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr11_+_327171 | 0.30 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chrX_+_49832231 | 0.30 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr7_+_100273736 | 0.29 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr20_+_32254286 | 0.29 |
ENST00000330271.4
|
ACTL10
|
actin-like 10 |
| chr19_-_50528584 | 0.29 |
ENST00000594092.1
ENST00000443401.2 ENST00000594948.1 ENST00000377011.2 ENST00000593919.1 ENST00000601324.1 ENST00000316763.3 ENST00000601341.1 ENST00000600259.1 |
VRK3
|
vaccinia related kinase 3 |
| chr7_-_37956409 | 0.29 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr19_-_40919271 | 0.29 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr22_+_19118321 | 0.29 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr10_+_134210672 | 0.29 |
ENST00000305233.5
ENST00000368609.4 |
PWWP2B
|
PWWP domain containing 2B |
| chr11_+_58938903 | 0.29 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr16_-_4588822 | 0.29 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr14_+_74003818 | 0.29 |
ENST00000311148.4
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr14_-_69261310 | 0.29 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr19_+_36119929 | 0.29 |
ENST00000588161.1
ENST00000262633.4 ENST00000592202.1 ENST00000586618.1 |
RBM42
|
RNA binding motif protein 42 |
| chr3_+_150126101 | 0.28 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chrX_-_48755030 | 0.28 |
ENST00000490755.2
ENST00000465150.2 ENST00000495490.2 |
TIMM17B
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr12_+_116985896 | 0.28 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr17_+_7155819 | 0.28 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr9_-_130477912 | 0.28 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr17_-_7835228 | 0.28 |
ENST00000303731.4
ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1
|
trafficking protein particle complex 1 |
| chr1_-_76076793 | 0.28 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr20_+_42295745 | 0.28 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr17_-_80231557 | 0.28 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr6_-_16761678 | 0.28 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr1_-_17231271 | 0.27 |
ENST00000606899.1
|
RP11-108M9.6
|
RP11-108M9.6 |
| chr17_+_38497640 | 0.27 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr19_-_51014588 | 0.27 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr12_-_28125638 | 0.27 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chrX_+_133941250 | 0.27 |
ENST00000445123.1
|
FAM122C
|
family with sequence similarity 122C |
| chr2_-_145188137 | 0.27 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr19_+_36120009 | 0.27 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr16_+_70380825 | 0.27 |
ENST00000417604.2
|
DDX19A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A |
| chrX_-_48824793 | 0.27 |
ENST00000376477.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr17_-_7155274 | 0.27 |
ENST00000318988.6
ENST00000575783.1 ENST00000573600.1 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr6_+_26204825 | 0.27 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr3_-_134092561 | 0.27 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr11_-_85376121 | 0.26 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr16_-_4588469 | 0.26 |
ENST00000588381.1
ENST00000563332.2 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr7_+_4721885 | 0.26 |
ENST00000328914.4
|
FOXK1
|
forkhead box K1 |
| chr21_-_44299626 | 0.26 |
ENST00000330317.2
ENST00000398208.2 |
WDR4
|
WD repeat domain 4 |
| chr17_+_29815113 | 0.26 |
ENST00000583755.1
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr19_+_49122548 | 0.26 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr16_+_31044413 | 0.26 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr9_+_131644398 | 0.26 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr20_-_1306351 | 0.26 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr9_+_131644388 | 0.26 |
ENST00000372600.4
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr15_+_68871308 | 0.26 |
ENST00000261861.5
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr22_-_38713394 | 0.26 |
ENST00000396832.1
ENST00000413574.2 |
CSNK1E
|
casein kinase 1, epsilon |
| chr19_+_57874835 | 0.26 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr11_+_818902 | 0.26 |
ENST00000336615.4
|
PNPLA2
|
patatin-like phospholipase domain containing 2 |
| chr10_+_123923205 | 0.25 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr12_+_6833323 | 0.25 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr11_-_116968987 | 0.25 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr19_-_44259136 | 0.25 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr14_-_75389925 | 0.25 |
ENST00000556776.1
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1 |
| chr17_-_7155802 | 0.25 |
ENST00000572043.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr19_+_36103631 | 0.25 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr16_+_29911666 | 0.25 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr16_-_11723066 | 0.25 |
ENST00000576036.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr22_-_38669030 | 0.25 |
ENST00000361906.3
|
TMEM184B
|
transmembrane protein 184B |
| chr7_+_100464760 | 0.25 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr4_-_7873981 | 0.25 |
ENST00000360265.4
|
AFAP1
|
actin filament associated protein 1 |
| chr17_-_7155775 | 0.25 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr14_+_96722152 | 0.25 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr19_+_8455077 | 0.25 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr2_-_220264703 | 0.24 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr19_-_53141584 | 0.24 |
ENST00000597161.1
ENST00000596930.1 ENST00000545872.1 ENST00000544146.1 ENST00000536937.1 ENST00000301096.3 |
ZNF83
|
zinc finger protein 83 |
| chr19_-_52227221 | 0.24 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr22_+_38597889 | 0.24 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr17_+_40610862 | 0.24 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr1_-_1293904 | 0.24 |
ENST00000309212.6
ENST00000342753.4 ENST00000445648.2 |
MXRA8
|
matrix-remodelling associated 8 |
| chrX_+_152338301 | 0.24 |
ENST00000453825.2
|
PNMA6A
|
paraneoplastic Ma antigen family member 6A |
| chr22_-_38713428 | 0.24 |
ENST00000403904.1
ENST00000405675.3 |
CSNK1E
|
casein kinase 1, epsilon |
| chr22_-_26879734 | 0.24 |
ENST00000422379.2
ENST00000336873.5 ENST00000398145.2 |
HPS4
|
Hermansky-Pudlak syndrome 4 |
| chr7_+_99905325 | 0.24 |
ENST00000332397.6
ENST00000437326.2 |
SPDYE3
|
speedy/RINGO cell cycle regulator family member E3 |
| chr11_-_65430554 | 0.24 |
ENST00000308639.9
ENST00000406246.3 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr2_+_238395042 | 0.24 |
ENST00000429898.1
ENST00000410032.1 |
MLPH
|
melanophilin |
| chr19_+_49467232 | 0.24 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr11_+_66610883 | 0.23 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr17_-_72864739 | 0.23 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr17_+_73089382 | 0.23 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr21_+_44589118 | 0.23 |
ENST00000291554.2
|
CRYAA
|
crystallin, alpha A |
| chr11_-_65430251 | 0.23 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chrX_+_133941218 | 0.23 |
ENST00000370784.4
ENST00000370785.3 |
FAM122C
|
family with sequence similarity 122C |
| chr8_-_41522779 | 0.23 |
ENST00000522231.1
ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1
|
ankyrin 1, erythrocytic |
| chr13_-_52378231 | 0.23 |
ENST00000280056.2
ENST00000444610.2 |
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr3_+_119814070 | 0.23 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr1_+_36023370 | 0.23 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr12_+_131438443 | 0.23 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr12_+_48876275 | 0.23 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr9_+_131644781 | 0.22 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of JUN
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 0.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 0.5 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.2 | 0.5 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.1 | 0.4 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.7 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.4 | GO:1990709 | maintenance of synapse structure(GO:0099558) presynaptic active zone organization(GO:1990709) |
| 0.1 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.3 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.1 | 0.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 1.6 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.3 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.1 | 0.2 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.3 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.9 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.5 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.7 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.3 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.3 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.1 | 0.3 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.4 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.3 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.3 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.2 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.3 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.4 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.7 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.4 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.5 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.4 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.5 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.4 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.3 | GO:1902748 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.9 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.8 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:1904744 | positive regulation of telomeric DNA binding(GO:1904744) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.5 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.4 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) convergent extension involved in organogenesis(GO:0060029) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.6 | GO:0051197 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.3 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.7 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.5 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.3 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 1.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.1 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) equilibrioception(GO:0050957) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 0.8 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 1.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.4 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.4 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 0.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 1.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.2 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 1.6 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.2 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0051427 | hormone receptor binding(GO:0051427) peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |