Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
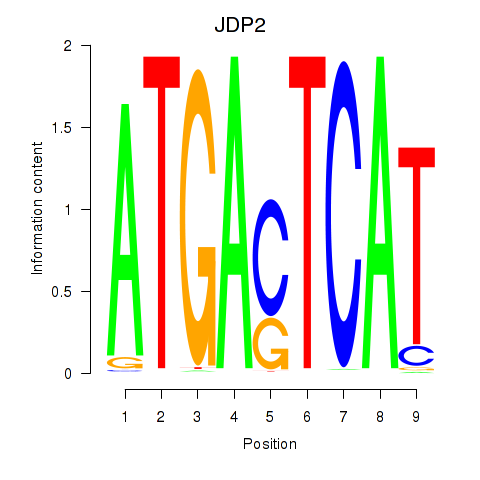
Results for JDP2
Z-value: 0.71
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JDP2 | hg19_v2_chr14_+_75894714_75894749 | -0.64 | 1.7e-01 | Click! |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_55162360 | 0.51 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr11_+_58874658 | 0.43 |
ENST00000411426.1
|
FAM111B
|
family with sequence similarity 111, member B |
| chr10_+_1102303 | 0.42 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr11_+_58874701 | 0.40 |
ENST00000529618.1
ENST00000534403.1 ENST00000343597.3 |
FAM111B
|
family with sequence similarity 111, member B |
| chr19_-_19739321 | 0.39 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr12_+_124997766 | 0.38 |
ENST00000543970.1
|
RP11-83B20.1
|
RP11-83B20.1 |
| chr19_-_35992780 | 0.37 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr10_-_105238997 | 0.36 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chr6_-_41673552 | 0.35 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr2_+_208104497 | 0.33 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr2_+_208104351 | 0.33 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr3_+_48507210 | 0.32 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr10_+_88854926 | 0.31 |
ENST00000298784.1
ENST00000298786.4 |
FAM35A
|
family with sequence similarity 35, member A |
| chr11_-_82782861 | 0.31 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr6_+_151358048 | 0.30 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr14_-_38028689 | 0.30 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chrY_+_15016725 | 0.29 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr5_+_169758393 | 0.29 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr8_+_40018977 | 0.28 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr15_+_57211318 | 0.27 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr11_-_8739566 | 0.26 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr8_+_22435762 | 0.26 |
ENST00000456545.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr1_+_17634689 | 0.26 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr7_+_30960915 | 0.26 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr2_+_202122826 | 0.25 |
ENST00000413726.1
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr11_-_2924720 | 0.24 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr12_-_105352080 | 0.24 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr3_-_98241760 | 0.23 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr1_-_108231101 | 0.23 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr4_-_146019693 | 0.23 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr5_-_157161727 | 0.23 |
ENST00000599823.1
|
AC026407.1
|
Uncharacterized protein |
| chr2_-_121624973 | 0.22 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chr12_-_10324716 | 0.22 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr6_+_292051 | 0.21 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr2_-_145275228 | 0.21 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_+_28724129 | 0.20 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr10_-_75385711 | 0.20 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr3_+_177545563 | 0.19 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr14_-_75083313 | 0.19 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr13_-_45048386 | 0.19 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr12_-_76817036 | 0.19 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr4_+_26578293 | 0.19 |
ENST00000512840.1
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr10_+_94594351 | 0.18 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr11_+_82783097 | 0.18 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr3_-_98241713 | 0.18 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chr19_+_38880695 | 0.18 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr20_-_48782639 | 0.18 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr1_-_204121013 | 0.18 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr2_+_87755054 | 0.18 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr7_+_1126437 | 0.17 |
ENST00000413368.1
ENST00000397092.1 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr3_+_138153451 | 0.17 |
ENST00000389567.4
ENST00000289135.4 |
ESYT3
|
extended synaptotagmin-like protein 3 |
| chr1_+_198189921 | 0.17 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr11_-_10920714 | 0.16 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr3_-_98241358 | 0.16 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr1_-_45272951 | 0.16 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr1_-_204121102 | 0.16 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr12_-_75784669 | 0.16 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr1_-_204121298 | 0.16 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr2_-_145275211 | 0.15 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_15114191 | 0.15 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr5_-_75919217 | 0.15 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr2_-_145275109 | 0.15 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr14_+_103801140 | 0.15 |
ENST00000561325.1
ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr3_-_48601206 | 0.15 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr6_-_113953705 | 0.14 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr4_+_86525299 | 0.14 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr11_-_57417367 | 0.14 |
ENST00000534810.1
|
YPEL4
|
yippee-like 4 (Drosophila) |
| chr7_-_135433460 | 0.14 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chrX_+_1734051 | 0.14 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr3_+_48507621 | 0.14 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr21_-_36421401 | 0.14 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr14_+_32476072 | 0.14 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr17_-_1508379 | 0.13 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr19_-_56047661 | 0.13 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr17_-_1553346 | 0.13 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr2_-_220173685 | 0.13 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr16_+_28996114 | 0.12 |
ENST00000395461.3
|
LAT
|
linker for activation of T cells |
| chr16_+_9449445 | 0.12 |
ENST00000564305.1
|
RP11-243A14.1
|
RP11-243A14.1 |
| chr3_-_98241598 | 0.12 |
ENST00000513287.1
ENST00000514537.1 ENST00000508071.1 ENST00000507944.1 |
CLDND1
|
claudin domain containing 1 |
| chr11_-_2924970 | 0.12 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr16_+_31366455 | 0.12 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr10_+_88428370 | 0.12 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr6_-_44400720 | 0.12 |
ENST00000595057.1
|
AL133262.1
|
AL133262.1 |
| chr8_-_123139423 | 0.11 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr12_+_75784850 | 0.11 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr5_-_67730240 | 0.11 |
ENST00000507733.1
|
CTC-537E7.3
|
CTC-537E7.3 |
| chr12_+_52695617 | 0.11 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr3_+_98699880 | 0.11 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr4_-_987217 | 0.11 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr2_+_46926048 | 0.11 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr15_+_89182178 | 0.11 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr20_+_4667094 | 0.11 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr9_+_138453595 | 0.11 |
ENST00000479141.1
ENST00000371766.2 ENST00000277508.5 ENST00000433563.1 |
PAEP
|
progestagen-associated endometrial protein |
| chr3_+_32023232 | 0.11 |
ENST00000360311.4
|
ZNF860
|
zinc finger protein 860 |
| chr11_+_67250490 | 0.10 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr7_-_142511084 | 0.10 |
ENST00000417977.2
|
TRBV30
|
T cell receptor beta variable 30 (gene/pseudogene) |
| chr3_+_28283069 | 0.10 |
ENST00000466830.1
ENST00000423894.1 |
CMC1
|
C-x(9)-C motif containing 1 |
| chr13_-_36429763 | 0.10 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr11_-_102401469 | 0.10 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr5_+_177540444 | 0.10 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chr19_+_50706866 | 0.10 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr11_-_8739383 | 0.10 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr5_-_149492904 | 0.10 |
ENST00000286301.3
ENST00000511344.1 |
CSF1R
|
colony stimulating factor 1 receptor |
| chr2_+_47596634 | 0.10 |
ENST00000419334.1
|
EPCAM
|
epithelial cell adhesion molecule |
| chr7_-_47492782 | 0.09 |
ENST00000413551.1
|
TNS3
|
tensin 3 |
| chr17_-_53800217 | 0.09 |
ENST00000424486.2
|
TMEM100
|
transmembrane protein 100 |
| chr16_+_31366536 | 0.09 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr10_-_21463116 | 0.09 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr9_+_34958254 | 0.09 |
ENST00000242315.3
|
KIAA1045
|
KIAA1045 |
| chr1_-_67142710 | 0.09 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr13_+_32838801 | 0.09 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr6_+_63921351 | 0.08 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr1_-_229230644 | 0.08 |
ENST00000433734.1
|
RP5-1065P14.2
|
RP5-1065P14.2 |
| chr9_+_44867571 | 0.08 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr6_+_47624172 | 0.08 |
ENST00000507065.1
ENST00000296862.1 |
GPR111
|
G protein-coupled receptor 111 |
| chr11_-_102714534 | 0.08 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr5_+_72143988 | 0.08 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr19_-_56056888 | 0.08 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chrX_-_15288154 | 0.08 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr16_+_28996364 | 0.08 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr5_-_148929848 | 0.07 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr3_-_151034734 | 0.07 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr1_-_244006528 | 0.07 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr7_-_155604967 | 0.07 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr17_+_74381343 | 0.07 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr5_+_135394840 | 0.07 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr20_-_17539456 | 0.07 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr16_-_14109841 | 0.07 |
ENST00000576797.1
ENST00000575424.1 |
CTD-2135D7.5
|
CTD-2135D7.5 |
| chr18_+_74207477 | 0.07 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr3_-_114343768 | 0.07 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_+_19988885 | 0.06 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr14_+_86401039 | 0.06 |
ENST00000557195.1
|
CTD-2341M24.1
|
CTD-2341M24.1 |
| chr15_-_54025300 | 0.06 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr17_+_66539369 | 0.06 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr17_+_30771279 | 0.06 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr11_-_82782952 | 0.06 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr12_-_102591604 | 0.06 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr1_-_166944652 | 0.06 |
ENST00000528703.1
ENST00000525740.1 ENST00000529387.1 ENST00000469934.2 ENST00000529071.1 ENST00000526687.1 |
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr3_-_149293990 | 0.06 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_+_35851570 | 0.05 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr12_-_131200810 | 0.05 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chr2_+_87754989 | 0.05 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr20_+_4666882 | 0.05 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr11_-_66103867 | 0.05 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr12_-_48152853 | 0.05 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr4_+_74606223 | 0.05 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr6_+_151042224 | 0.05 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chrX_+_47053208 | 0.05 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr2_-_99870744 | 0.05 |
ENST00000409238.1
ENST00000423800.1 |
LYG2
|
lysozyme G-like 2 |
| chr9_-_124991124 | 0.05 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr11_-_119599794 | 0.05 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr12_+_75760714 | 0.05 |
ENST00000547144.1
|
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr5_+_145583156 | 0.05 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr7_-_57207571 | 0.05 |
ENST00000331162.4
|
ZNF479
|
zinc finger protein 479 |
| chr10_-_4285923 | 0.04 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr11_+_6220376 | 0.04 |
ENST00000311352.2
|
OR52W1
|
olfactory receptor, family 52, subfamily W, member 1 |
| chr17_+_28256874 | 0.04 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr12_+_6644443 | 0.04 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chrX_+_133371077 | 0.04 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr3_-_18466787 | 0.04 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr5_+_145583107 | 0.04 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr21_+_26934165 | 0.04 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr10_-_31288398 | 0.04 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr17_-_38956205 | 0.04 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr2_+_169926047 | 0.04 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr10_+_52152766 | 0.04 |
ENST00000596442.1
|
AC069547.2
|
Uncharacterized protein |
| chr1_-_160231451 | 0.04 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr1_+_152881014 | 0.04 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr1_-_153521714 | 0.04 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr19_+_49467232 | 0.04 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr6_-_49755019 | 0.04 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr9_-_21335240 | 0.04 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chr3_-_45957088 | 0.04 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr6_-_31745037 | 0.04 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr6_-_31745085 | 0.04 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
| chr1_+_151227179 | 0.03 |
ENST00000368884.3
ENST00000368881.4 |
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr4_+_84457529 | 0.03 |
ENST00000264409.4
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr21_-_36421626 | 0.03 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr6_+_125540951 | 0.03 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chrX_-_154493791 | 0.03 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr5_+_176237478 | 0.03 |
ENST00000329542.4
|
UNC5A
|
unc-5 homolog A (C. elegans) |
| chr16_+_57662138 | 0.03 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr7_-_135433534 | 0.03 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chr2_+_68872954 | 0.03 |
ENST00000394342.2
|
PROKR1
|
prokineticin receptor 1 |
| chr11_-_75201791 | 0.03 |
ENST00000529721.1
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr16_+_2083265 | 0.03 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr11_+_57308979 | 0.03 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr14_+_73525144 | 0.03 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr17_+_79650962 | 0.03 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr3_+_69811858 | 0.03 |
ENST00000433517.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr9_-_23826298 | 0.03 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr21_-_36421535 | 0.03 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr16_-_4164027 | 0.03 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr20_+_42136308 | 0.03 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr4_+_84457250 | 0.03 |
ENST00000395226.2
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr8_-_38386175 | 0.03 |
ENST00000437935.2
ENST00000358138.1 |
C8orf86
|
chromosome 8 open reading frame 86 |
| chr8_-_27472198 | 0.03 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chr19_-_51872233 | 0.03 |
ENST00000601435.1
ENST00000291715.1 |
CLDND2
|
claudin domain containing 2 |
| chr7_+_48128316 | 0.03 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr5_-_41794663 | 0.02 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030185 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 | 0.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.2 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.1 | 0.5 | GO:1902748 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.2 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0030805 | regulation of cyclic nucleotide catabolic process(GO:0030805) positive regulation of cyclic nucleotide catabolic process(GO:0030807) regulation of cAMP catabolic process(GO:0030820) positive regulation of cAMP catabolic process(GO:0030822) regulation of purine nucleotide catabolic process(GO:0033121) positive regulation of purine nucleotide catabolic process(GO:0033123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |