Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
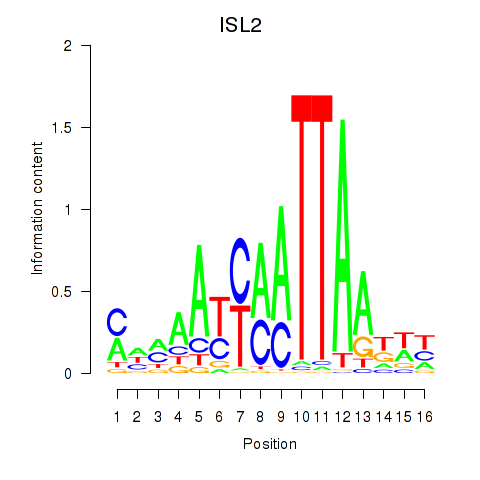
Results for ISL2
Z-value: 0.69
Transcription factors associated with ISL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL2
|
ENSG00000159556.5 | ISL LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL2 | hg19_v2_chr15_+_76629064_76629073 | -0.03 | 9.5e-01 | Click! |
Activity profile of ISL2 motif
Sorted Z-values of ISL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_4710288 | 0.71 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr14_+_61449197 | 0.59 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr4_+_95128748 | 0.54 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr10_+_63422695 | 0.43 |
ENST00000330194.2
ENST00000389639.3 |
C10orf107
|
chromosome 10 open reading frame 107 |
| chr7_-_81635106 | 0.37 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr4_+_169013666 | 0.36 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr5_+_40841410 | 0.35 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr12_+_75784850 | 0.32 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr3_+_172468505 | 0.30 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr14_-_51027838 | 0.29 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr3_+_172468472 | 0.28 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr3_+_136649311 | 0.28 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr5_-_54988448 | 0.27 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr14_+_57671888 | 0.26 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr18_-_64271316 | 0.26 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chrX_-_84634737 | 0.25 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr9_-_99540328 | 0.24 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr11_-_58378759 | 0.24 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr16_+_53133070 | 0.22 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr18_-_53303123 | 0.22 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr4_+_128702969 | 0.21 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr1_+_152178320 | 0.21 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr11_-_28129656 | 0.21 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr4_+_25162253 | 0.21 |
ENST00000512921.1
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chrM_+_10053 | 0.21 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr5_+_32788945 | 0.21 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr15_-_98417780 | 0.21 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr12_+_64798095 | 0.20 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr2_-_87248975 | 0.20 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chrX_+_108779004 | 0.19 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr10_-_115904361 | 0.19 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chrM_+_9207 | 0.18 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr12_-_58212487 | 0.18 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr6_-_111804905 | 0.18 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr4_+_26344754 | 0.17 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr5_+_110427983 | 0.17 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr4_+_95128996 | 0.17 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr16_-_3350614 | 0.17 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr3_-_20227619 | 0.16 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr11_+_28129795 | 0.15 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr14_-_73997901 | 0.15 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr4_-_185275104 | 0.15 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr14_+_45605157 | 0.15 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr16_-_1538765 | 0.15 |
ENST00000447419.2
ENST00000440447.2 |
PTX4
|
pentraxin 4, long |
| chr15_+_90735145 | 0.15 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr3_+_138327542 | 0.14 |
ENST00000360570.3
ENST00000393035.2 |
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_99127225 | 0.14 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr18_+_61575200 | 0.14 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr8_-_121457608 | 0.14 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr3_+_138327417 | 0.14 |
ENST00000338446.4
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_+_156775910 | 0.14 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chrX_-_21676442 | 0.14 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr5_-_54988559 | 0.13 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr6_+_64345698 | 0.13 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr14_+_61449076 | 0.13 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr5_-_130500922 | 0.13 |
ENST00000513012.1
ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr3_-_183145873 | 0.13 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr2_+_86669118 | 0.12 |
ENST00000427678.1
ENST00000542128.1 |
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr2_-_190446738 | 0.12 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr1_-_223853348 | 0.12 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr13_+_24144796 | 0.12 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr12_+_122356488 | 0.11 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr2_+_32502952 | 0.11 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chrX_-_84634708 | 0.11 |
ENST00000373145.3
|
POF1B
|
premature ovarian failure, 1B |
| chrX_+_96138907 | 0.11 |
ENST00000373040.3
|
RPA4
|
replication protein A4, 30kDa |
| chr4_+_26324474 | 0.11 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_-_110937351 | 0.11 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr1_-_238108575 | 0.11 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr2_+_105050794 | 0.10 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr11_+_73498898 | 0.10 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr3_-_193096600 | 0.10 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr18_-_64271363 | 0.10 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr1_-_152779104 | 0.09 |
ENST00000606576.1
ENST00000368768.1 |
LCE1C
|
late cornified envelope 1C |
| chr21_-_30445886 | 0.09 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr2_+_231921574 | 0.09 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr3_-_141747950 | 0.09 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_113568207 | 0.09 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_-_228613026 | 0.09 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr14_-_53258314 | 0.09 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr12_+_64798826 | 0.09 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr14_-_60043524 | 0.09 |
ENST00000537690.2
ENST00000281581.4 |
CCDC175
|
coiled-coil domain containing 175 |
| chr13_+_24144509 | 0.09 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_-_74025625 | 0.09 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr12_-_54689532 | 0.08 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr6_-_24358264 | 0.08 |
ENST00000378454.3
|
DCDC2
|
doublecortin domain containing 2 |
| chr9_+_35042205 | 0.08 |
ENST00000312292.5
ENST00000378745.3 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr2_+_234668894 | 0.08 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr14_+_102276132 | 0.08 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr13_+_49551020 | 0.08 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr13_+_78315295 | 0.08 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr6_+_26440700 | 0.08 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr7_-_16844611 | 0.08 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr11_-_107729287 | 0.08 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr21_+_15588499 | 0.07 |
ENST00000400577.3
|
RBM11
|
RNA binding motif protein 11 |
| chrX_-_55208866 | 0.07 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr11_-_107729504 | 0.07 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr17_-_64225508 | 0.07 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr10_-_27529486 | 0.07 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr13_+_78315348 | 0.07 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_-_121740969 | 0.07 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr1_-_93257951 | 0.06 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr14_-_55907334 | 0.06 |
ENST00000247219.5
|
TBPL2
|
TATA box binding protein like 2 |
| chr12_-_10282742 | 0.06 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr12_+_26164645 | 0.06 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr2_+_88047606 | 0.06 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr1_-_168464875 | 0.06 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr13_-_20806440 | 0.05 |
ENST00000400066.3
ENST00000400065.3 ENST00000356192.6 |
GJB6
|
gap junction protein, beta 6, 30kDa |
| chr1_+_171227069 | 0.05 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr19_+_29456034 | 0.05 |
ENST00000589921.1
|
LINC00906
|
long intergenic non-protein coding RNA 906 |
| chr4_-_120243545 | 0.05 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_-_40680578 | 0.05 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr7_-_37488777 | 0.05 |
ENST00000445322.1
ENST00000448602.1 |
ELMO1
|
engulfment and cell motility 1 |
| chr12_+_104337515 | 0.05 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr3_-_157221380 | 0.04 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr15_-_83378638 | 0.04 |
ENST00000261722.3
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr11_-_63376013 | 0.04 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr3_+_134514093 | 0.04 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr3_-_72150076 | 0.04 |
ENST00000488545.1
ENST00000608654.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr2_-_231860596 | 0.03 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr5_+_68860949 | 0.03 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr4_-_69111401 | 0.03 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr2_-_136633940 | 0.03 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr7_+_13141010 | 0.03 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr5_+_40909354 | 0.03 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr2_-_217559517 | 0.02 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr11_-_327537 | 0.02 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr11_-_92931098 | 0.02 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chrX_-_32173579 | 0.02 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr14_-_23058063 | 0.02 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.4 |
DAD1
|
defender against cell death 1 |
| chr17_-_33390667 | 0.02 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr6_+_130339710 | 0.02 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr5_+_40841276 | 0.02 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr2_+_234580525 | 0.02 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_66820058 | 0.01 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_+_59194154 | 0.01 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr20_-_50722183 | 0.01 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr5_-_41794313 | 0.01 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr1_+_234765057 | 0.01 |
ENST00000429269.1
|
LINC00184
|
long intergenic non-protein coding RNA 184 |
| chr3_+_139063372 | 0.00 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr2_+_234580499 | 0.00 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr7_-_37488834 | 0.00 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr19_-_51920952 | 0.00 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr2_-_207078154 | 0.00 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr19_-_36001113 | 0.00 |
ENST00000434389.1
|
DMKN
|
dermokine |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.4 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.1 | GO:0060345 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.3 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0046292 | spermatid nucleus elongation(GO:0007290) formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:1903899 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |