Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
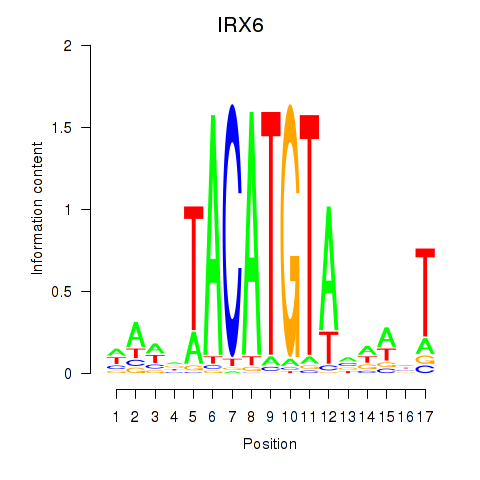
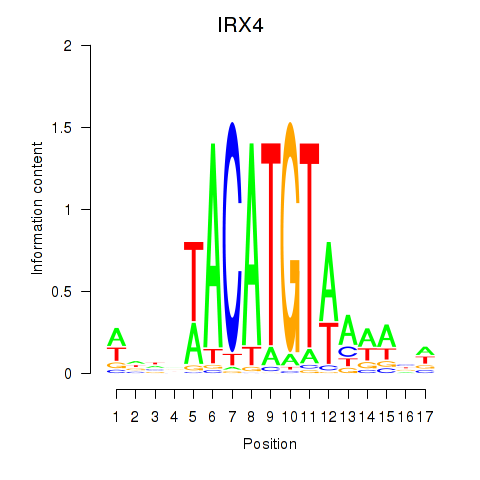
Results for IRX6_IRX4
Z-value: 0.26
Transcription factors associated with IRX6_IRX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX6
|
ENSG00000159387.7 | iroquois homeobox 6 |
|
IRX4
|
ENSG00000113430.5 | iroquois homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX4 | hg19_v2_chr5_-_1882858_1883003 | -0.41 | 4.2e-01 | Click! |
| IRX6 | hg19_v2_chr16_+_55357672_55357672 | -0.10 | 8.5e-01 | Click! |
Activity profile of IRX6_IRX4 motif
Sorted Z-values of IRX6_IRX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_141264597 | 0.62 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr12_-_123728548 | 0.17 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr9_+_26746951 | 0.17 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr3_+_53528659 | 0.16 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr4_-_104119528 | 0.14 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr3_-_27498235 | 0.12 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr12_-_76462713 | 0.11 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr7_+_23210760 | 0.11 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr15_-_63448973 | 0.09 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr6_+_149887377 | 0.09 |
ENST00000367419.5
|
GINM1
|
glycoprotein integral membrane 1 |
| chr4_-_104119488 | 0.09 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr1_+_63989004 | 0.08 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr4_+_130017268 | 0.08 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr8_+_84824920 | 0.07 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr3_+_186158169 | 0.07 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr7_-_16844611 | 0.07 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr15_-_64665911 | 0.07 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr17_+_38673270 | 0.07 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr2_+_160590469 | 0.06 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr4_+_123653882 | 0.05 |
ENST00000433287.1
|
BBS12
|
Bardet-Biedl syndrome 12 |
| chr12_+_32655048 | 0.05 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr8_+_27631903 | 0.05 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_+_220267429 | 0.05 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr14_+_56127960 | 0.04 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr14_+_56127989 | 0.04 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr8_-_17555164 | 0.04 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr2_+_58655520 | 0.04 |
ENST00000455219.3
ENST00000449448.2 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr4_-_143481822 | 0.04 |
ENST00000510812.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr8_+_67976593 | 0.04 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr17_-_29641104 | 0.04 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr1_+_212475148 | 0.04 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr6_-_33679452 | 0.04 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr22_-_24096562 | 0.03 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr15_+_81225699 | 0.03 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr10_+_90660832 | 0.03 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr20_+_10199468 | 0.03 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr2_+_58655461 | 0.03 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr10_-_115904361 | 0.03 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chrX_-_19988382 | 0.03 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr2_-_238333919 | 0.03 |
ENST00000409162.1
|
AC112721.1
|
Uncharacterized protein |
| chr4_+_113558612 | 0.02 |
ENST00000505034.1
ENST00000324052.6 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr6_-_117747015 | 0.02 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr3_-_139199589 | 0.02 |
ENST00000506825.1
|
RBP2
|
retinol binding protein 2, cellular |
| chr18_-_46784778 | 0.02 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr18_-_28622774 | 0.02 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr18_-_47376197 | 0.02 |
ENST00000592688.1
|
MYO5B
|
myosin VB |
| chr13_-_41768654 | 0.02 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr3_-_168865522 | 0.02 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr7_+_6121296 | 0.02 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr8_-_67976509 | 0.02 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr4_+_158141899 | 0.02 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr10_+_86184676 | 0.01 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr1_-_152086556 | 0.01 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr2_-_231989808 | 0.01 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr4_+_158142750 | 0.01 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_-_153303658 | 0.01 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr3_+_111630451 | 0.01 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr2_+_113670548 | 0.01 |
ENST00000263326.3
ENST00000352179.3 ENST00000349806.3 ENST00000353225.3 |
IL37
|
interleukin 37 |
| chr9_-_27573651 | 0.01 |
ENST00000379995.1
ENST00000379997.3 |
C9orf72
|
chromosome 9 open reading frame 72 |
| chr18_-_70305745 | 0.01 |
ENST00000581073.1
|
CBLN2
|
cerebellin 2 precursor |
| chr1_+_53308398 | 0.01 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chrX_+_99899180 | 0.01 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr12_+_32655110 | 0.01 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr16_+_777118 | 0.01 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr14_-_106845789 | 0.00 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr4_+_41361616 | 0.00 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_+_111758553 | 0.00 |
ENST00000419127.1
|
TMPRSS7
|
transmembrane protease, serine 7 |
| chr21_-_48024986 | 0.00 |
ENST00000291700.4
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chr1_+_67673297 | 0.00 |
ENST00000425614.1
ENST00000395227.1 |
IL23R
|
interleukin 23 receptor |
| chr3_+_171561127 | 0.00 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr14_-_68159152 | 0.00 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr15_+_66585555 | 0.00 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX6_IRX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |