Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
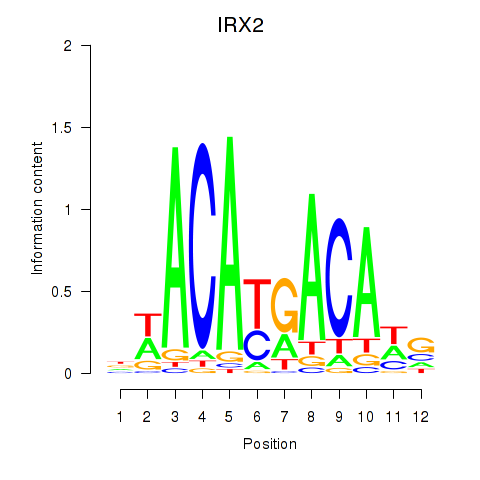
Results for IRX2
Z-value: 0.46
Transcription factors associated with IRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX2
|
ENSG00000170561.8 | iroquois homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX2 | hg19_v2_chr5_-_2751762_2751784 | 0.72 | 1.1e-01 | Click! |
Activity profile of IRX2 motif
Sorted Z-values of IRX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_148855726 | 0.21 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X linked 1 |
| chrX_-_148676974 | 0.21 |
ENST00000524178.1
|
HSFX2
|
heat shock transcription factor family, X linked 2 |
| chr1_-_8939265 | 0.18 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr22_+_45714672 | 0.18 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr4_+_22999152 | 0.17 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr6_+_13272904 | 0.16 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr4_-_83821676 | 0.16 |
ENST00000355196.2
ENST00000507676.1 ENST00000506495.1 ENST00000507051.1 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr17_+_76037081 | 0.16 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr9_+_130159593 | 0.14 |
ENST00000419132.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr1_-_151148442 | 0.13 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chrX_-_70331298 | 0.12 |
ENST00000456850.2
ENST00000473378.1 ENST00000487883.1 ENST00000374202.2 |
IL2RG
|
interleukin 2 receptor, gamma |
| chr2_+_159651821 | 0.12 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr1_-_151148492 | 0.12 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr2_-_46844242 | 0.12 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr7_-_105332084 | 0.12 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr11_+_33037652 | 0.12 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr14_+_52327350 | 0.11 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr13_+_35516390 | 0.11 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr4_+_88896819 | 0.11 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr19_+_13134772 | 0.11 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_-_238649319 | 0.10 |
ENST00000400946.2
|
RP11-371I1.2
|
long intergenic non-protein coding RNA 1139 |
| chr3_-_186262166 | 0.10 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chrX_+_155227246 | 0.10 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr20_+_2083540 | 0.10 |
ENST00000400064.3
|
STK35
|
serine/threonine kinase 35 |
| chr13_-_24007815 | 0.10 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr3_-_185641681 | 0.10 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chrX_-_100129128 | 0.10 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr8_-_90993869 | 0.09 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr1_-_149783914 | 0.09 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr11_+_77532233 | 0.09 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr1_+_244227632 | 0.09 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr9_-_35812236 | 0.09 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr16_+_58537737 | 0.09 |
ENST00000561738.1
|
NDRG4
|
NDRG family member 4 |
| chr2_-_3504587 | 0.08 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr11_+_35222629 | 0.08 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr16_+_21244986 | 0.08 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr8_+_26240414 | 0.08 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr20_-_35329063 | 0.08 |
ENST00000422536.1
|
NDRG3
|
NDRG family member 3 |
| chr11_-_118305921 | 0.08 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr2_-_42180940 | 0.07 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr10_-_4285923 | 0.07 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr3_-_11645925 | 0.07 |
ENST00000413604.1
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr17_-_41322332 | 0.07 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr12_+_4714145 | 0.07 |
ENST00000545342.1
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr3_+_193853927 | 0.07 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chrX_+_155227371 | 0.07 |
ENST00000369423.2
ENST00000540897.1 |
IL9R
|
interleukin 9 receptor |
| chr11_+_62648357 | 0.07 |
ENST00000541372.1
ENST00000539458.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr10_+_70320413 | 0.06 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr5_-_71803177 | 0.06 |
ENST00000414109.2
|
ZNF366
|
zinc finger protein 366 |
| chr5_-_150138246 | 0.06 |
ENST00000518015.1
|
DCTN4
|
dynactin 4 (p62) |
| chr8_-_124665190 | 0.06 |
ENST00000325995.7
|
KLHL38
|
kelch-like family member 38 |
| chr8_+_67104323 | 0.06 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr12_+_53773944 | 0.05 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr21_-_22175341 | 0.05 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr7_-_33140498 | 0.05 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr11_-_33795893 | 0.05 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr7_+_93652116 | 0.05 |
ENST00000415536.1
|
AC003092.1
|
AC003092.1 |
| chrX_-_100129320 | 0.05 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr2_+_46844290 | 0.05 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr13_-_46679144 | 0.05 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr3_-_114343768 | 0.05 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr20_-_60294804 | 0.05 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr11_+_62648336 | 0.05 |
ENST00000338663.7
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr7_+_150065278 | 0.05 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr10_+_46997926 | 0.05 |
ENST00000374314.4
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr2_-_98280383 | 0.05 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr17_+_34842512 | 0.05 |
ENST00000588253.1
ENST00000592616.1 ENST00000590858.1 ENST00000588357.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr1_-_111506562 | 0.05 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr5_-_138210977 | 0.05 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr7_-_130598059 | 0.05 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr3_+_184534994 | 0.05 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr17_-_7761172 | 0.05 |
ENST00000333775.5
ENST00000575771.1 |
LSMD1
|
LSM domain containing 1 |
| chr17_+_7761013 | 0.05 |
ENST00000571846.1
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr1_-_173793458 | 0.04 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr2_-_70520539 | 0.04 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr4_-_80247162 | 0.04 |
ENST00000286794.4
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr16_+_57680043 | 0.04 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr14_+_52327109 | 0.04 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr8_+_67405794 | 0.04 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr8_+_98900132 | 0.04 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr1_+_113010056 | 0.04 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr2_-_166060552 | 0.04 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr7_-_130597935 | 0.04 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr9_-_79267432 | 0.04 |
ENST00000424866.1
|
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr1_+_67673297 | 0.04 |
ENST00000425614.1
ENST00000395227.1 |
IL23R
|
interleukin 23 receptor |
| chr9_+_79792410 | 0.04 |
ENST00000357409.5
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chrX_-_21676442 | 0.04 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr10_+_102672712 | 0.04 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr6_-_53013620 | 0.04 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr5_-_150138061 | 0.04 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr14_+_62462541 | 0.04 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr3_-_126327398 | 0.04 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr16_-_12062333 | 0.03 |
ENST00000597717.1
|
AC007216.2
|
Uncharacterized protein |
| chr19_+_3762645 | 0.03 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr2_+_196313239 | 0.03 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr2_+_27851863 | 0.03 |
ENST00000264718.3
ENST00000610189.1 |
GPN1
|
GPN-loop GTPase 1 |
| chr17_-_8055747 | 0.03 |
ENST00000317276.4
ENST00000581703.1 |
PER1
|
period circadian clock 1 |
| chr11_-_76381029 | 0.03 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr20_+_56136136 | 0.03 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr17_-_60885645 | 0.03 |
ENST00000544856.2
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr2_+_171640291 | 0.03 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr18_-_53070913 | 0.03 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr9_+_12695702 | 0.03 |
ENST00000381136.2
|
TYRP1
|
tyrosinase-related protein 1 |
| chr1_+_113009163 | 0.03 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr15_-_44486632 | 0.03 |
ENST00000484674.1
|
FRMD5
|
FERM domain containing 5 |
| chr3_+_148457585 | 0.03 |
ENST00000402260.1
|
AGTR1
|
angiotensin II receptor, type 1 |
| chr4_-_153332886 | 0.03 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr13_+_28519343 | 0.03 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr16_+_56899114 | 0.03 |
ENST00000566786.1
ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr1_+_17516275 | 0.03 |
ENST00000412427.1
|
RP11-380J14.1
|
RP11-380J14.1 |
| chr3_-_194072019 | 0.03 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr9_-_47314222 | 0.02 |
ENST00000420228.1
ENST00000438517.1 ENST00000414020.1 |
AL953854.2
|
AL953854.2 |
| chr17_+_34842473 | 0.02 |
ENST00000490126.2
ENST00000225410.4 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr19_-_40336969 | 0.02 |
ENST00000599134.1
ENST00000597634.1 ENST00000598417.1 ENST00000601274.1 ENST00000594309.1 ENST00000221801.3 |
FBL
|
fibrillarin |
| chr12_-_66275350 | 0.02 |
ENST00000536648.1
|
RP11-366L20.2
|
Uncharacterized protein |
| chr17_+_900342 | 0.02 |
ENST00000327158.4
|
TIMM22
|
translocase of inner mitochondrial membrane 22 homolog (yeast) |
| chr16_+_33205585 | 0.02 |
ENST00000360260.2
ENST00000398666.3 |
TP53TG3C
|
TP53 target 3C |
| chr16_+_33262120 | 0.02 |
ENST00000569741.1
ENST00000380147.3 |
TP53TG3B
|
TP53 target 3B |
| chr15_+_44580955 | 0.02 |
ENST00000345795.2
ENST00000360824.3 |
CASC4
|
cancer susceptibility candidate 4 |
| chr12_-_112856623 | 0.02 |
ENST00000551291.2
|
RPL6
|
ribosomal protein L6 |
| chr8_+_24151553 | 0.02 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr17_-_7761256 | 0.02 |
ENST00000575208.1
|
LSMD1
|
LSM domain containing 1 |
| chr5_-_138211051 | 0.02 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr4_+_109571740 | 0.02 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr8_-_11996586 | 0.02 |
ENST00000333796.3
|
USP17L2
|
ubiquitin specific peptidase 17-like family member 2 |
| chr10_+_96522361 | 0.02 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr1_-_190446759 | 0.02 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr14_-_64971893 | 0.02 |
ENST00000555220.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr12_+_9144626 | 0.02 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr14_+_58797974 | 0.02 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_+_95115210 | 0.02 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr10_+_90660832 | 0.02 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr16_-_32687431 | 0.02 |
ENST00000398680.3
ENST00000569420.1 |
TP53TG3
|
TP53 target 3 |
| chr8_-_29939933 | 0.02 |
ENST00000522794.1
|
TMEM66
|
transmembrane protein 66 |
| chr6_-_27841289 | 0.02 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr2_+_171034646 | 0.02 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr12_-_16758304 | 0.02 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr14_+_23790655 | 0.01 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr4_+_158142750 | 0.01 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr2_-_46844159 | 0.01 |
ENST00000474980.1
ENST00000306465.4 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr12_+_90313394 | 0.01 |
ENST00000549551.1
|
RP11-654D12.2
|
RP11-654D12.2 |
| chr12_+_48178706 | 0.01 |
ENST00000599515.1
|
AC004466.1
|
Uncharacterized protein |
| chr10_-_100027943 | 0.01 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr4_+_158141899 | 0.01 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr9_-_73736511 | 0.01 |
ENST00000377110.3
ENST00000377111.2 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr3_-_150612089 | 0.01 |
ENST00000397891.3
|
FAM188B2
|
family with sequence similarity 188, member B2 |
| chr4_+_119606523 | 0.01 |
ENST00000388822.5
ENST00000506780.1 ENST00000508801.1 |
METTL14
|
methyltransferase like 14 |
| chr12_-_91505608 | 0.01 |
ENST00000266718.4
|
LUM
|
lumican |
| chr4_-_168155417 | 0.01 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr13_-_22033392 | 0.01 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr2_-_166060571 | 0.01 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr3_-_58419537 | 0.01 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr7_-_14880892 | 0.01 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr17_-_60885700 | 0.01 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr21_+_43823983 | 0.01 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr18_+_50278430 | 0.01 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr9_-_23779367 | 0.01 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr5_-_94417186 | 0.01 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr4_+_159131596 | 0.01 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr2_+_168675182 | 0.01 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr17_-_27054952 | 0.01 |
ENST00000580518.1
|
TLCD1
|
TLC domain containing 1 |
| chr4_+_158141843 | 0.01 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr17_-_60885659 | 0.01 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr14_+_75536280 | 0.00 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr11_-_110561721 | 0.00 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr2_+_133874577 | 0.00 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr3_+_46616017 | 0.00 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr16_-_4401258 | 0.00 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr17_-_2614927 | 0.00 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr14_+_75536335 | 0.00 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr1_+_204839959 | 0.00 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr12_+_54402790 | 0.00 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr12_+_128399965 | 0.00 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr8_+_81397876 | 0.00 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr9_+_34957477 | 0.00 |
ENST00000544237.1
|
KIAA1045
|
KIAA1045 |
| chr16_-_71323617 | 0.00 |
ENST00000563876.1
|
CMTR2
|
cap methyltransferase 2 |
| chr3_+_31574189 | 0.00 |
ENST00000295770.2
|
STT3B
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr2_-_233877912 | 0.00 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr8_-_82359662 | 0.00 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr2_-_166060382 | 0.00 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr12_+_128399917 | 0.00 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.2 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.0 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |