Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
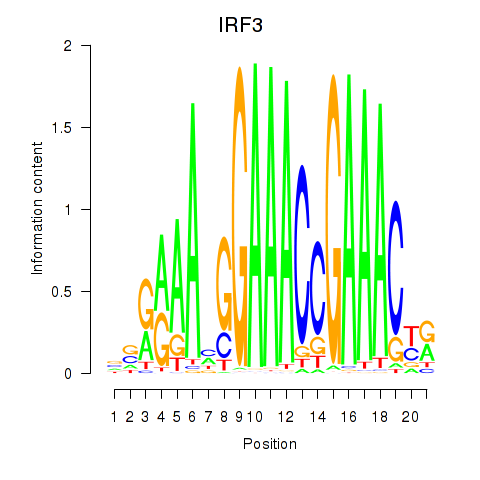
Results for IRF3
Z-value: 4.12
Transcription factors associated with IRF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF3
|
ENSG00000126456.11 | interferon regulatory factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF3 | hg19_v2_chr19_-_50169064_50169132 | 0.30 | 5.6e-01 | Click! |
Activity profile of IRF3 motif
Sorted Z-values of IRF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_948803 | 34.40 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr4_-_169239921 | 17.56 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr21_+_42792442 | 16.63 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr11_-_615570 | 14.18 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr11_-_615942 | 13.66 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr10_+_91087651 | 13.31 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr2_-_163175133 | 12.93 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr10_+_91092241 | 11.34 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr1_+_79115503 | 9.10 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr3_-_122283079 | 9.00 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_122283100 | 8.67 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_122283424 | 8.43 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr7_-_92747269 | 7.98 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chr12_+_113376157 | 6.54 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr12_+_113376249 | 6.24 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr19_-_17516449 | 6.06 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr12_+_113416191 | 5.53 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr9_-_32526184 | 5.14 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr2_-_231084659 | 5.09 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr3_+_122283175 | 5.05 |
ENST00000383661.3
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr9_-_32526299 | 4.98 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr3_+_122283064 | 4.92 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr7_-_92777606 | 4.79 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr2_-_231084820 | 4.44 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr12_+_113416265 | 4.38 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr2_-_37384175 | 3.72 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr3_+_122399444 | 3.23 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr10_+_91061712 | 3.14 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr4_-_169401628 | 3.13 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr11_+_5646213 | 3.09 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr2_-_231084617 | 2.79 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr22_+_36649170 | 2.75 |
ENST00000438034.1
ENST00000427990.1 ENST00000347595.7 ENST00000397279.4 ENST00000433768.1 ENST00000440669.2 |
APOL1
|
apolipoprotein L, 1 |
| chr19_+_10196781 | 2.72 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr4_+_89378261 | 2.71 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr7_-_139756791 | 2.45 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr11_-_57334732 | 2.22 |
ENST00000526659.1
ENST00000527022.1 |
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr14_+_24605389 | 2.21 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr3_+_159706537 | 2.20 |
ENST00000305579.2
ENST00000480787.1 ENST00000466512.1 |
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr19_+_10196981 | 2.19 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr12_+_133614062 | 2.17 |
ENST00000540031.1
ENST00000536123.1 |
ZNF84
|
zinc finger protein 84 |
| chr19_+_49977466 | 2.03 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr22_+_18632666 | 1.92 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr22_+_36649056 | 1.91 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
| chr11_-_57335280 | 1.85 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr14_+_24605361 | 1.81 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr12_+_113416340 | 1.79 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr8_+_26150628 | 1.74 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr7_-_944631 | 1.74 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr12_+_133613937 | 1.56 |
ENST00000539354.1
ENST00000542874.1 ENST00000438628.2 |
ZNF84
|
zinc finger protein 84 |
| chr1_-_154580616 | 1.41 |
ENST00000368474.4
|
ADAR
|
adenosine deaminase, RNA-specific |
| chr3_+_122399697 | 1.37 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr4_-_74847800 | 1.33 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
| chr17_+_8316442 | 1.30 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr20_-_47894569 | 1.27 |
ENST00000371744.1
ENST00000371752.1 ENST00000396105.1 |
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr14_-_24615523 | 1.22 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr2_-_99485825 | 1.20 |
ENST00000423771.1
|
KIAA1211L
|
KIAA1211-like |
| chr17_-_40264321 | 1.20 |
ENST00000430773.1
ENST00000413196.2 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr3_-_123339343 | 1.13 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr4_+_130017268 | 1.13 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr12_+_133613878 | 1.10 |
ENST00000392319.2
ENST00000543758.1 |
ZNF84
|
zinc finger protein 84 |
| chr17_-_7166500 | 1.08 |
ENST00000575313.1
ENST00000397317.4 |
CLDN7
|
claudin 7 |
| chr1_+_76190357 | 1.05 |
ENST00000370834.5
ENST00000541113.1 ENST00000543667.1 ENST00000420607.2 |
ACADM
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr22_+_36044411 | 1.04 |
ENST00000409652.4
|
APOL6
|
apolipoprotein L, 6 |
| chr14_-_24615805 | 0.96 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr12_-_10022735 | 0.95 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr10_+_35416223 | 0.95 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr17_-_40264692 | 0.95 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr10_+_35415978 | 0.93 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr1_+_25598872 | 0.92 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr11_-_4414880 | 0.92 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr9_-_100954910 | 0.85 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr8_-_67525473 | 0.84 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr6_-_36355513 | 0.79 |
ENST00000340181.4
ENST00000373737.4 |
ETV7
|
ets variant 7 |
| chr15_-_55611306 | 0.77 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr6_-_2876744 | 0.77 |
ENST00000420981.2
|
RP11-420G6.4
|
RP11-420G6.4 |
| chr10_-_70231639 | 0.76 |
ENST00000551118.2
ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2
|
DNA replication helicase/nuclease 2 |
| chr14_+_73525229 | 0.75 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr17_-_30669138 | 0.73 |
ENST00000225805.4
ENST00000577809.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr2_+_99771418 | 0.72 |
ENST00000393473.2
ENST00000393477.3 ENST00000393474.3 ENST00000340066.1 ENST00000393471.2 ENST00000449211.1 ENST00000434566.1 ENST00000410042.1 |
LIPT1
MRPL30
|
lipoyltransferase 1 39S ribosomal protein L30, mitochondrial |
| chr2_+_69240302 | 0.72 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr6_-_27840099 | 0.70 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr5_-_96143796 | 0.69 |
ENST00000296754.3
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr3_+_184529948 | 0.67 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr1_-_177939041 | 0.67 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr15_+_45003675 | 0.66 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr3_+_184529929 | 0.63 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr12_+_133614119 | 0.62 |
ENST00000327668.7
|
ZNF84
|
zinc finger protein 84 |
| chr14_+_73525144 | 0.61 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr14_-_91976874 | 0.60 |
ENST00000557018.1
|
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr4_-_87855851 | 0.59 |
ENST00000473559.1
|
C4orf36
|
chromosome 4 open reading frame 36 |
| chr9_-_95087838 | 0.54 |
ENST00000442668.2
ENST00000421075.2 ENST00000536624.1 |
NOL8
|
nucleolar protein 8 |
| chrX_+_119384607 | 0.53 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
| chr12_+_97300995 | 0.53 |
ENST00000266742.4
ENST00000429527.2 ENST00000554226.1 ENST00000557478.1 ENST00000557092.1 ENST00000411739.2 ENST00000553609.1 |
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr6_+_26440700 | 0.51 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr6_-_41039567 | 0.51 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_+_207139367 | 0.50 |
ENST00000374423.3
|
ZDBF2
|
zinc finger, DBF-type containing 2 |
| chr11_+_118230287 | 0.48 |
ENST00000252108.3
ENST00000431736.2 |
UBE4A
|
ubiquitination factor E4A |
| chr11_+_125461607 | 0.46 |
ENST00000527606.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr15_+_55611128 | 0.46 |
ENST00000164305.5
ENST00000539642.1 |
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr16_-_71323271 | 0.45 |
ENST00000565850.1
ENST00000568910.1 ENST00000434935.2 ENST00000338099.5 |
CMTR2
|
cap methyltransferase 2 |
| chr5_+_77656339 | 0.43 |
ENST00000538629.1
|
SCAMP1
|
secretory carrier membrane protein 1 |
| chr6_-_36355486 | 0.42 |
ENST00000538992.1
|
ETV7
|
ets variant 7 |
| chr7_-_138794394 | 0.42 |
ENST00000242351.5
ENST00000471652.1 |
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr10_-_14996321 | 0.41 |
ENST00000378289.4
|
DCLRE1C
|
DNA cross-link repair 1C |
| chr17_-_63052929 | 0.40 |
ENST00000439174.2
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13 |
| chr14_+_86401039 | 0.39 |
ENST00000557195.1
|
CTD-2341M24.1
|
CTD-2341M24.1 |
| chr16_-_28223229 | 0.39 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
| chr6_-_28226984 | 0.39 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr7_-_138794081 | 0.37 |
ENST00000464606.1
|
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr9_-_95087604 | 0.36 |
ENST00000542613.1
ENST00000542053.1 ENST00000358855.4 ENST00000545558.1 ENST00000432670.2 ENST00000433029.2 ENST00000411621.2 |
NOL8
|
nucleolar protein 8 |
| chr7_-_32931387 | 0.36 |
ENST00000304056.4
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr1_-_177939348 | 0.35 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr2_-_214017151 | 0.34 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr19_+_13051206 | 0.34 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr6_+_32821924 | 0.33 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_-_121655593 | 0.33 |
ENST00000398212.2
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr2_-_152118352 | 0.32 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr14_+_35452104 | 0.30 |
ENST00000216774.6
ENST00000546080.1 |
SRP54
|
signal recognition particle 54kDa |
| chr20_+_46988646 | 0.30 |
ENST00000416742.1
ENST00000425021.1 |
LINC00494
|
long intergenic non-protein coding RNA 494 |
| chr6_-_32806506 | 0.30 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr8_+_22446763 | 0.28 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr9_+_74764340 | 0.28 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr9_+_5450503 | 0.28 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr17_-_61517572 | 0.26 |
ENST00000582997.1
|
CYB561
|
cytochrome b561 |
| chr6_-_32821599 | 0.26 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr1_-_89664595 | 0.25 |
ENST00000355754.6
|
GBP4
|
guanylate binding protein 4 |
| chr2_+_69240415 | 0.25 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr1_+_112298514 | 0.23 |
ENST00000536167.1
|
DDX20
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 |
| chr14_+_35452169 | 0.22 |
ENST00000555557.1
|
SRP54
|
signal recognition particle 54kDa |
| chrX_+_146993449 | 0.22 |
ENST00000218200.8
ENST00000370471.3 ENST00000370477.1 |
FMR1
|
fragile X mental retardation 1 |
| chr15_+_55611401 | 0.22 |
ENST00000566999.1
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr5_-_96143602 | 0.22 |
ENST00000443439.2
ENST00000503921.1 ENST00000508227.1 ENST00000507154.1 |
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr4_-_156875003 | 0.22 |
ENST00000433477.3
|
CTSO
|
cathepsin O |
| chr22_-_36635684 | 0.18 |
ENST00000358502.5
|
APOL2
|
apolipoprotein L, 2 |
| chr1_-_146644122 | 0.18 |
ENST00000254101.3
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr12_-_322821 | 0.17 |
ENST00000359674.4
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr16_+_75681650 | 0.16 |
ENST00000300086.4
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr20_-_47894936 | 0.15 |
ENST00000371754.4
|
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr4_+_124320665 | 0.15 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr17_+_6659153 | 0.14 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr12_-_133613794 | 0.12 |
ENST00000443154.3
|
RP11-386I8.6
|
RP11-386I8.6 |
| chrX_+_77154935 | 0.11 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr3_-_125802998 | 0.09 |
ENST00000514677.1
|
SLC41A3
|
solute carrier family 41, member 3 |
| chrX_-_109561294 | 0.08 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr7_-_126892303 | 0.07 |
ENST00000358373.3
|
GRM8
|
glutamate receptor, metabotropic 8 |
| chr2_+_219125714 | 0.07 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr6_+_28193037 | 0.06 |
ENST00000531981.1
ENST00000425468.2 ENST00000252207.5 ENST00000531979.1 ENST00000527436.1 |
ZSCAN9
|
zinc finger and SCAN domain containing 9 |
| chr3_-_160283348 | 0.06 |
ENST00000334256.4
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3) |
| chr9_-_95640218 | 0.06 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr7_-_37382360 | 0.06 |
ENST00000455119.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr17_+_21729899 | 0.05 |
ENST00000583708.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr5_+_169758393 | 0.05 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr6_-_110964453 | 0.04 |
ENST00000413605.2
|
CDK19
|
cyclin-dependent kinase 19 |
| chr3_+_51977294 | 0.03 |
ENST00000498510.1
|
PARP3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr5_-_180288248 | 0.03 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr12_-_8218997 | 0.02 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr1_-_11042094 | 0.02 |
ENST00000377004.4
ENST00000377008.4 |
C1orf127
|
chromosome 1 open reading frame 127 |
| chrX_+_146993534 | 0.02 |
ENST00000334557.6
ENST00000439526.2 ENST00000370475.4 |
FMR1
|
fragile X mental retardation 1 |
| chr2_-_190044480 | 0.01 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr6_-_32806483 | 0.01 |
ENST00000374899.4
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr4_+_144434584 | 0.01 |
ENST00000283131.3
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr14_+_73525265 | 0.01 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 27.8 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 5.8 | 23.0 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 3.4 | 41.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 3.3 | 19.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 2.4 | 26.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 2.1 | 27.8 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 2.0 | 6.1 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 1.1 | 12.8 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 1.1 | 11.7 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.9 | 14.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.8 | 16.6 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.5 | 1.4 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.4 | 3.7 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.3 | 1.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 1.0 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.3 | 2.3 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 | 1.0 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.2 | 0.9 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 | 1.3 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.2 | 1.5 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) regulation of osteoclast proliferation(GO:0090289) |
| 0.2 | 10.0 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.2 | 0.8 | GO:0043504 | DNA replication, removal of RNA primer(GO:0043137) mitochondrial DNA repair(GO:0043504) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.4 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 4.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 1.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.5 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 1.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.9 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 1.9 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 0.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 4.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 3.1 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 1.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 1.3 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 1.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 1.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 2.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.2 | 4.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.7 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 1.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.9 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 6.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 16.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 11.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 8.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 32.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 21.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 29.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 34.4 | GO:0031386 | protein tag(GO:0031386) |
| 2.0 | 24.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.9 | 26.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.8 | 6.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 6.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.6 | 3.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.4 | 1.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 42.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.3 | 1.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.3 | 2.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 1.0 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.2 | 0.9 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.2 | 7.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 27.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 6.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.7 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.3 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.6 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) TAP1 binding(GO:0046978) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 16.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 4.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 10.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 3.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 27.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 3.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 50.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 1.1 | 98.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 6.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 5.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 3.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 6.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |