Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
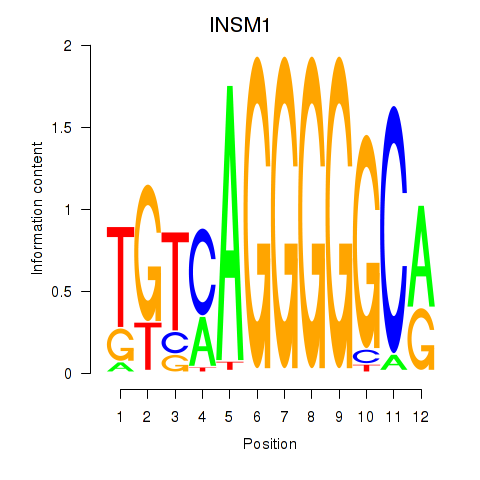
Results for INSM1
Z-value: 0.63
Transcription factors associated with INSM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
INSM1
|
ENSG00000173404.3 | INSM transcriptional repressor 1 |
Activity profile of INSM1 motif
Sorted Z-values of INSM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_10197463 | 0.62 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr4_-_164253738 | 0.57 |
ENST00000509586.1
ENST00000504391.1 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr3_+_50192457 | 0.46 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr10_+_35416223 | 0.37 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr15_+_89182178 | 0.34 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr10_+_91061712 | 0.34 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr19_+_10196781 | 0.34 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr14_+_65879668 | 0.33 |
ENST00000553924.1
ENST00000358307.2 ENST00000557338.1 ENST00000554610.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr19_+_55795493 | 0.31 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr3_+_50192499 | 0.31 |
ENST00000413852.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr15_+_89181974 | 0.30 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr14_-_62162541 | 0.28 |
ENST00000557544.1
|
HIF1A-AS1
|
HIF1A antisense RNA 1 |
| chr15_+_45722727 | 0.28 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr19_+_10196981 | 0.28 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_+_153191706 | 0.27 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr4_-_129209221 | 0.27 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr15_+_89182156 | 0.27 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr3_+_50192537 | 0.25 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr14_+_31343951 | 0.24 |
ENST00000556908.1
ENST00000555881.1 ENST00000460581.2 |
COCH
|
cochlin |
| chr14_+_31343747 | 0.23 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr12_-_80328700 | 0.22 |
ENST00000550107.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr13_-_52027134 | 0.22 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chrX_+_16804544 | 0.22 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr15_-_52263937 | 0.21 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr14_+_65879437 | 0.21 |
ENST00000394585.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr2_-_153573887 | 0.20 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr4_+_174089904 | 0.20 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr4_+_26322409 | 0.19 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_-_147443043 | 0.19 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr17_-_4889508 | 0.19 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr14_+_62162258 | 0.18 |
ENST00000337138.4
ENST00000394997.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr19_+_49977466 | 0.18 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr13_-_36705425 | 0.18 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr9_+_131452239 | 0.18 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr1_-_236445251 | 0.18 |
ENST00000354619.5
ENST00000327333.8 |
ERO1LB
|
ERO1-like beta (S. cerevisiae) |
| chr13_-_31736132 | 0.17 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr8_-_103136481 | 0.17 |
ENST00000524209.1
ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD
|
neurocalcin delta |
| chr17_-_36413133 | 0.16 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr17_-_42992856 | 0.16 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr5_+_154237778 | 0.16 |
ENST00000523698.1
ENST00000517876.1 ENST00000520472.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr15_+_79166065 | 0.15 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr6_+_155054459 | 0.15 |
ENST00000367178.3
ENST00000417268.1 ENST00000367186.4 |
SCAF8
|
SR-related CTD-associated factor 8 |
| chr1_+_65613340 | 0.15 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr7_-_91509986 | 0.15 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr5_+_154238042 | 0.14 |
ENST00000519211.1
ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chrX_+_118892545 | 0.14 |
ENST00000343905.3
|
SOWAHD
|
sosondowah ankyrin repeat domain family member D |
| chr10_+_35415851 | 0.14 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chrX_-_71525742 | 0.14 |
ENST00000450875.1
ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr6_-_97345689 | 0.13 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr12_-_51420108 | 0.13 |
ENST00000547198.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr16_+_610407 | 0.13 |
ENST00000409413.3
|
C16orf11
|
chromosome 16 open reading frame 11 |
| chr6_+_52442083 | 0.13 |
ENST00000606714.1
|
TRAM2-AS1
|
TRAM2 antisense RNA 1 (head to head) |
| chr11_+_10772847 | 0.12 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr11_-_46142615 | 0.12 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr7_+_94536898 | 0.12 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr6_+_56820018 | 0.12 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr1_+_65613217 | 0.12 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr7_-_105752651 | 0.12 |
ENST00000470347.1
ENST00000455385.2 |
SYPL1
|
synaptophysin-like 1 |
| chr12_-_52887034 | 0.12 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr10_+_134258649 | 0.11 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chr5_-_141030943 | 0.11 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr8_-_124286495 | 0.11 |
ENST00000297857.2
|
ZHX1
|
zinc fingers and homeoboxes 1 |
| chr20_-_45980621 | 0.11 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr4_-_147442817 | 0.11 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr5_+_154238096 | 0.11 |
ENST00000517568.1
ENST00000524105.1 ENST00000285896.6 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr22_+_20104947 | 0.11 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr18_+_34409069 | 0.11 |
ENST00000543923.1
ENST00000280020.5 ENST00000435985.2 ENST00000592521.1 ENST00000587139.1 |
KIAA1328
|
KIAA1328 |
| chr3_-_48601206 | 0.11 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr1_+_168148273 | 0.10 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr9_-_26892342 | 0.10 |
ENST00000535437.1
|
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr13_-_52026730 | 0.10 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr17_+_45608430 | 0.10 |
ENST00000322157.4
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr5_+_154238149 | 0.10 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr12_+_49372251 | 0.10 |
ENST00000293549.3
|
WNT1
|
wingless-type MMTV integration site family, member 1 |
| chr1_+_160370344 | 0.10 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr4_-_129208940 | 0.10 |
ENST00000296425.5
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr12_-_51419924 | 0.10 |
ENST00000541174.2
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr17_+_52978185 | 0.10 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr19_-_55881741 | 0.10 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr16_-_30134524 | 0.10 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr9_-_112083229 | 0.10 |
ENST00000374566.3
ENST00000374557.4 |
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B |
| chr17_-_42441204 | 0.10 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chrX_+_69674943 | 0.10 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr12_-_118810688 | 0.10 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chrX_-_153095813 | 0.10 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr12_-_51420128 | 0.09 |
ENST00000262051.7
ENST00000547732.1 ENST00000262052.5 ENST00000546488.1 ENST00000550714.1 ENST00000548193.1 ENST00000547579.1 ENST00000546743.1 |
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr10_+_88516396 | 0.09 |
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA |
| chr5_-_171711061 | 0.09 |
ENST00000393792.2
|
UBTD2
|
ubiquitin domain containing 2 |
| chr17_-_56606705 | 0.09 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr1_+_10490441 | 0.09 |
ENST00000470413.2
ENST00000309048.3 |
APITD1-CORT
APITD1
|
APITD1-CORT readthrough apoptosis-inducing, TAF9-like domain 1 |
| chr2_+_24272543 | 0.09 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr1_-_205325850 | 0.09 |
ENST00000537168.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr16_+_88704978 | 0.09 |
ENST00000244241.4
|
IL17C
|
interleukin 17C |
| chr16_-_30134441 | 0.09 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr15_+_27216530 | 0.09 |
ENST00000555083.1
|
GABRG3
|
gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
| chr8_-_67525524 | 0.08 |
ENST00000517885.1
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr12_-_76953513 | 0.08 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr3_+_46924829 | 0.08 |
ENST00000313049.5
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr9_-_130517522 | 0.08 |
ENST00000373274.3
ENST00000420366.1 |
SH2D3C
|
SH2 domain containing 3C |
| chr2_+_152266604 | 0.08 |
ENST00000430328.2
|
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr7_-_105752971 | 0.08 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chr17_+_34058639 | 0.08 |
ENST00000268864.3
|
RASL10B
|
RAS-like, family 10, member B |
| chr1_+_153750622 | 0.08 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr20_-_30310797 | 0.08 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr17_+_52978156 | 0.08 |
ENST00000348161.4
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr15_-_51058005 | 0.08 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr12_-_50101003 | 0.08 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr5_-_60140009 | 0.08 |
ENST00000505959.1
|
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr17_-_56606664 | 0.07 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr1_-_154531095 | 0.07 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr1_-_205325698 | 0.07 |
ENST00000460687.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr17_+_45608614 | 0.07 |
ENST00000544660.1
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr13_-_31736027 | 0.07 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_+_44679370 | 0.07 |
ENST00000372290.4
|
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr8_-_67525473 | 0.07 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr2_+_207139367 | 0.06 |
ENST00000374423.3
|
ZDBF2
|
zinc finger, DBF-type containing 2 |
| chr11_-_47399942 | 0.06 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr17_-_56606639 | 0.06 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr9_-_98269481 | 0.06 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr17_+_52978107 | 0.06 |
ENST00000445275.2
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr11_-_47400062 | 0.06 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr17_+_27369918 | 0.06 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr8_+_99076509 | 0.06 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr6_-_17706618 | 0.06 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr2_+_152266392 | 0.06 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chrX_-_41782249 | 0.06 |
ENST00000442742.2
ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr10_+_35415978 | 0.06 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr7_-_132262060 | 0.06 |
ENST00000359827.3
|
PLXNA4
|
plexin A4 |
| chr1_-_205326022 | 0.06 |
ENST00000367155.3
|
KLHDC8A
|
kelch domain containing 8A |
| chr11_-_47400078 | 0.06 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr15_-_75748143 | 0.05 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr22_+_45064593 | 0.05 |
ENST00000432186.1
|
PRR5
|
proline rich 5 (renal) |
| chr12_-_76953573 | 0.05 |
ENST00000549646.1
ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr15_-_43802769 | 0.05 |
ENST00000263801.3
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr11_+_58346584 | 0.05 |
ENST00000316059.6
|
ZFP91
|
ZFP91 zinc finger protein |
| chr1_-_235116495 | 0.05 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr6_+_56820152 | 0.05 |
ENST00000370745.1
|
BEND6
|
BEN domain containing 6 |
| chr12_-_54779511 | 0.05 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr1_-_6557156 | 0.05 |
ENST00000537245.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr9_+_115983808 | 0.05 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr2_+_24272576 | 0.05 |
ENST00000380986.4
ENST00000452109.1 |
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr17_-_36762095 | 0.05 |
ENST00000578925.1
ENST00000264659.7 |
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr19_+_42817450 | 0.05 |
ENST00000301204.3
|
TMEM145
|
transmembrane protein 145 |
| chr7_-_91509972 | 0.05 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr1_+_2398876 | 0.05 |
ENST00000449969.1
|
PLCH2
|
phospholipase C, eta 2 |
| chr15_+_27216297 | 0.05 |
ENST00000333743.6
|
GABRG3
|
gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
| chr1_+_156863470 | 0.05 |
ENST00000338302.3
ENST00000455314.1 ENST00000292357.7 |
PEAR1
|
platelet endothelial aggregation receptor 1 |
| chr4_+_174089951 | 0.04 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr11_+_10772534 | 0.04 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr6_-_146285221 | 0.04 |
ENST00000367503.3
ENST00000438092.2 ENST00000275233.7 |
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr20_-_45981138 | 0.04 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr1_-_205326161 | 0.04 |
ENST00000367156.3
ENST00000606887.1 ENST00000607173.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr9_-_130517309 | 0.04 |
ENST00000414380.1
|
SH2D3C
|
SH2 domain containing 3C |
| chrX_+_49020121 | 0.04 |
ENST00000415364.1
ENST00000376338.3 ENST00000425285.1 |
MAGIX
|
MAGI family member, X-linked |
| chr20_+_44657845 | 0.04 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr9_-_80646374 | 0.04 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr2_+_208394794 | 0.04 |
ENST00000536726.1
ENST00000374397.4 ENST00000452474.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr2_+_232575128 | 0.04 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr1_-_205325994 | 0.04 |
ENST00000491471.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr17_-_42462688 | 0.04 |
ENST00000377068.3
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr3_+_97540876 | 0.04 |
ENST00000419587.2
|
CRYBG3
|
Beta/gamma crystallin domain-containing protein 3; cDNA FLJ60082, weakly similar to Uro-adherence factor A |
| chr7_-_84816122 | 0.04 |
ENST00000444867.1
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr11_+_120110863 | 0.04 |
ENST00000543440.2
|
POU2F3
|
POU class 2 homeobox 3 |
| chr19_+_35630022 | 0.03 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_-_37980344 | 0.03 |
ENST00000448519.2
ENST00000373075.2 ENST00000373073.4 ENST00000296214.5 |
MEAF6
|
MYST/Esa1-associated factor 6 |
| chr8_-_101571933 | 0.03 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr14_+_102027688 | 0.03 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr4_-_83931862 | 0.03 |
ENST00000506560.1
ENST00000442461.2 ENST00000446851.2 ENST00000340417.3 |
LIN54
|
lin-54 homolog (C. elegans) |
| chr1_+_54411715 | 0.03 |
ENST00000371370.3
ENST00000371368.1 |
LRRC42
|
leucine rich repeat containing 42 |
| chr4_-_143767428 | 0.03 |
ENST00000513000.1
ENST00000509777.1 ENST00000503927.1 |
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr9_+_136243264 | 0.03 |
ENST00000371955.1
|
C9orf96
|
chromosome 9 open reading frame 96 |
| chr14_+_76776957 | 0.03 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr11_-_46142948 | 0.03 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr9_+_37650945 | 0.03 |
ENST00000377765.3
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr17_-_47755436 | 0.03 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr4_-_151936865 | 0.03 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr10_+_115999004 | 0.03 |
ENST00000603594.1
|
VWA2
|
von Willebrand factor A domain containing 2 |
| chr18_-_34408693 | 0.03 |
ENST00000587382.1
ENST00000589049.1 ENST00000587129.1 |
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr5_-_60140089 | 0.03 |
ENST00000507047.1
ENST00000438340.1 ENST00000425382.1 ENST00000508821.1 |
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr6_+_56819773 | 0.03 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr11_-_57417367 | 0.03 |
ENST00000534810.1
|
YPEL4
|
yippee-like 4 (Drosophila) |
| chr17_-_73389854 | 0.03 |
ENST00000578961.1
ENST00000392564.1 ENST00000582582.1 |
GRB2
|
growth factor receptor-bound protein 2 |
| chr1_+_210001309 | 0.03 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr22_+_20105012 | 0.03 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr4_-_151936416 | 0.03 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr17_+_42081914 | 0.03 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr21_-_46707793 | 0.03 |
ENST00000331343.7
ENST00000349485.5 |
POFUT2
|
protein O-fucosyltransferase 2 |
| chr12_+_58148842 | 0.03 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr19_-_38916802 | 0.03 |
ENST00000587738.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr12_-_52715179 | 0.02 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr9_+_137979506 | 0.02 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr3_+_35680339 | 0.02 |
ENST00000450234.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_-_94129244 | 0.02 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr7_-_79082786 | 0.02 |
ENST00000522391.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr22_-_30695471 | 0.02 |
ENST00000434291.1
|
RP1-130H16.18
|
Uncharacterized protein |
| chr17_-_47755338 | 0.02 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr3_+_179040767 | 0.02 |
ENST00000496856.1
ENST00000491818.1 |
ZNF639
|
zinc finger protein 639 |
| chr2_-_73511559 | 0.02 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr1_-_85725316 | 0.02 |
ENST00000344356.5
ENST00000471115.1 |
C1orf52
|
chromosome 1 open reading frame 52 |
| chr6_-_146285455 | 0.02 |
ENST00000367505.2
|
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
Network of associatons between targets according to the STRING database.
First level regulatory network of INSM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.5 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.2 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.1 | 1.0 | GO:1901166 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.2 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.2 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 0.1 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.1 | GO:0061184 | Spemann organizer formation(GO:0060061) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.2 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.3 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0048372 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) regulation of osteoclast proliferation(GO:0090289) |
| 0.0 | 0.1 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.0 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.6 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.5 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.3 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |