Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
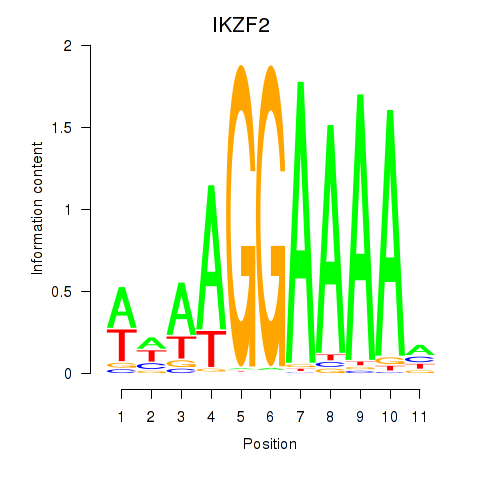
Results for IKZF2
Z-value: 0.48
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.12 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg19_v2_chr2_-_214015111_214015179 | 0.70 | 1.2e-01 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_17909594 | 0.63 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_+_97597148 | 0.32 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr9_+_92219919 | 0.31 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr6_-_44400720 | 0.26 |
ENST00000595057.1
|
AL133262.1
|
AL133262.1 |
| chr10_-_118897567 | 0.22 |
ENST00000369206.5
|
VAX1
|
ventral anterior homeobox 1 |
| chr12_+_50690489 | 0.21 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chrX_+_73164149 | 0.20 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr10_-_76868931 | 0.20 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr12_-_122240792 | 0.19 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr12_+_79371565 | 0.18 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr5_+_35852797 | 0.17 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr8_+_101349823 | 0.17 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chrX_+_78003204 | 0.17 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr4_-_185303418 | 0.16 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr5_+_32710736 | 0.16 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr10_-_10504285 | 0.16 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr5_-_68339648 | 0.15 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr12_+_28605426 | 0.15 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr8_+_39770803 | 0.15 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr15_+_69373210 | 0.15 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chrX_+_49832231 | 0.15 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr12_+_116985896 | 0.15 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr6_+_27107053 | 0.15 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr12_+_26348582 | 0.15 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr6_-_112081113 | 0.14 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr7_+_66800928 | 0.14 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr11_-_69867159 | 0.14 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr2_+_87808725 | 0.14 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr7_+_123469037 | 0.14 |
ENST00000489978.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr3_+_193853927 | 0.14 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr1_-_238649319 | 0.14 |
ENST00000400946.2
|
RP11-371I1.2
|
long intergenic non-protein coding RNA 1139 |
| chr5_-_42811986 | 0.14 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr2_+_27301435 | 0.13 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr17_-_6915616 | 0.13 |
ENST00000575889.1
|
AC027763.2
|
Uncharacterized protein |
| chr3_-_27764190 | 0.13 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr6_-_152639479 | 0.13 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr10_+_99079008 | 0.13 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr4_-_153303658 | 0.13 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_+_57138437 | 0.13 |
ENST00000504228.1
ENST00000541073.1 |
KIAA1211
|
KIAA1211 |
| chr19_+_50433476 | 0.13 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr12_-_28125638 | 0.13 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr16_+_50313426 | 0.13 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr14_+_85994943 | 0.12 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr8_+_77596014 | 0.12 |
ENST00000523885.1
|
ZFHX4
|
zinc finger homeobox 4 |
| chr4_-_798689 | 0.12 |
ENST00000504062.1
|
CPLX1
|
complexin 1 |
| chr8_-_123706338 | 0.12 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr10_-_75168071 | 0.12 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr14_+_91709103 | 0.12 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chrX_+_52841228 | 0.11 |
ENST00000351072.1
ENST00000425386.1 ENST00000375503.3 |
XAGE5
|
X antigen family, member 5 |
| chr7_+_73442487 | 0.11 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr2_-_214014959 | 0.11 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr17_+_7387919 | 0.11 |
ENST00000572844.1
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr12_-_91573132 | 0.11 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr3_-_128690173 | 0.11 |
ENST00000508239.1
|
RP11-723O4.6
|
Uncharacterized protein FLJ43738 |
| chr7_+_138482695 | 0.10 |
ENST00000422794.2
ENST00000397602.3 ENST00000442682.2 ENST00000458494.1 ENST00000413208.1 |
TMEM213
|
transmembrane protein 213 |
| chr20_-_43133491 | 0.10 |
ENST00000411544.1
|
SERINC3
|
serine incorporator 3 |
| chrX_-_15683147 | 0.10 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr1_+_104615595 | 0.10 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr16_+_20911978 | 0.10 |
ENST00000562740.1
|
LYRM1
|
LYR motif containing 1 |
| chr14_-_23479331 | 0.10 |
ENST00000397377.1
ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr8_-_36636676 | 0.10 |
ENST00000524132.1
ENST00000519451.1 |
RP11-962G15.1
|
RP11-962G15.1 |
| chr11_+_20044375 | 0.10 |
ENST00000525322.1
ENST00000530408.1 |
NAV2
|
neuron navigator 2 |
| chr17_+_7387677 | 0.10 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr22_-_39640756 | 0.09 |
ENST00000331163.6
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr1_+_95616933 | 0.09 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr2_+_232135245 | 0.09 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr1_+_117963209 | 0.09 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr2_+_98330009 | 0.09 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr1_-_151148492 | 0.09 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr6_+_148593425 | 0.09 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr12_-_71551652 | 0.09 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr1_+_174933899 | 0.09 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr22_+_38203898 | 0.09 |
ENST00000323205.6
ENST00000248924.6 ENST00000445195.1 |
GCAT
|
glycine C-acetyltransferase |
| chr6_+_26365443 | 0.09 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr6_-_10435032 | 0.09 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr8_+_144120648 | 0.09 |
ENST00000395172.1
|
C8orf31
|
chromosome 8 open reading frame 31 |
| chr1_+_111888890 | 0.09 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr16_+_20911174 | 0.09 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr21_+_17791838 | 0.09 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr15_-_75918787 | 0.09 |
ENST00000564086.1
|
SNUPN
|
snurportin 1 |
| chr12_-_56321397 | 0.09 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr1_+_36038971 | 0.09 |
ENST00000373235.3
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr9_+_132099158 | 0.09 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr5_+_80529104 | 0.09 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr11_+_20044096 | 0.09 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr4_-_168155577 | 0.08 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_+_81892477 | 0.08 |
ENST00000372263.3
|
PLAC9
|
placenta-specific 9 |
| chr1_-_157014865 | 0.08 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr19_+_24009879 | 0.08 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr6_+_32944119 | 0.08 |
ENST00000606059.1
|
BRD2
|
bromodomain containing 2 |
| chr6_+_36839616 | 0.08 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr9_+_5510492 | 0.08 |
ENST00000397745.2
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr4_-_146859623 | 0.08 |
ENST00000379448.4
ENST00000513320.1 |
ZNF827
|
zinc finger protein 827 |
| chr12_+_26348429 | 0.08 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr8_+_9046503 | 0.08 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr12_-_11287243 | 0.08 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr10_-_106240032 | 0.08 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr12_-_123921256 | 0.08 |
ENST00000280571.8
|
RILPL2
|
Rab interacting lysosomal protein-like 2 |
| chr17_-_19364269 | 0.08 |
ENST00000421796.2
ENST00000585389.1 ENST00000609249.1 |
AC004448.5
|
AC004448.5 |
| chr19_-_58071166 | 0.08 |
ENST00000601415.1
|
ZNF550
|
zinc finger protein 550 |
| chr15_+_40731920 | 0.08 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr12_+_52450298 | 0.08 |
ENST00000550582.2
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr17_-_6915646 | 0.08 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr4_-_185776854 | 0.08 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr6_+_147830063 | 0.08 |
ENST00000367474.1
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr15_-_82338460 | 0.07 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr19_-_47290535 | 0.07 |
ENST00000412532.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr11_-_67141090 | 0.07 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr6_-_26056695 | 0.07 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr16_+_89334512 | 0.07 |
ENST00000602042.1
|
AC137932.1
|
AC137932.1 |
| chr15_+_63050785 | 0.07 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr16_+_72459838 | 0.07 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chrX_-_63005405 | 0.07 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr3_-_71294304 | 0.07 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr16_-_28374829 | 0.07 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr6_+_106546808 | 0.07 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr12_+_8309630 | 0.07 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr2_+_20650796 | 0.07 |
ENST00000448241.1
|
AC023137.2
|
AC023137.2 |
| chr8_-_101724989 | 0.07 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_49449107 | 0.07 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr15_-_66545995 | 0.07 |
ENST00000395614.1
ENST00000288745.3 ENST00000422354.1 ENST00000395625.2 ENST00000360698.4 ENST00000409699.2 |
MEGF11
|
multiple EGF-like-domains 11 |
| chr18_+_22040593 | 0.07 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr17_+_35294075 | 0.07 |
ENST00000254457.5
|
LHX1
|
LIM homeobox 1 |
| chrX_+_149887090 | 0.06 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr16_+_75032901 | 0.06 |
ENST00000335325.4
ENST00000320619.6 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr21_+_17791648 | 0.06 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrX_-_74743080 | 0.06 |
ENST00000373367.3
|
ZDHHC15
|
zinc finger, DHHC-type containing 15 |
| chr12_+_26348246 | 0.06 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr3_-_151176497 | 0.06 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr16_+_81272287 | 0.06 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr11_+_208815 | 0.06 |
ENST00000530889.1
|
RIC8A
|
RIC8 guanine nucleotide exchange factor A |
| chr3_-_46068969 | 0.06 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr9_-_139334247 | 0.06 |
ENST00000371712.3
|
INPP5E
|
inositol polyphosphate-5-phosphatase, 72 kDa |
| chr17_-_17740325 | 0.06 |
ENST00000338854.5
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr7_+_134671234 | 0.06 |
ENST00000436302.2
ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein-like 3 |
| chr3_+_186435065 | 0.06 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr1_-_161337662 | 0.06 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr20_-_43589109 | 0.06 |
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr19_-_44031341 | 0.06 |
ENST00000600651.1
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr10_+_48189612 | 0.06 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr4_-_74486109 | 0.06 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr19_+_48773337 | 0.06 |
ENST00000595607.1
|
ZNF114
|
zinc finger protein 114 |
| chr2_-_214015111 | 0.06 |
ENST00000433134.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr9_+_5510558 | 0.06 |
ENST00000397747.3
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr15_-_75918500 | 0.06 |
ENST00000569817.1
|
SNUPN
|
snurportin 1 |
| chr22_+_40573921 | 0.06 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr10_+_97709725 | 0.06 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr6_+_106534192 | 0.06 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr17_-_66951382 | 0.06 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr18_+_50278430 | 0.06 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr12_-_68845165 | 0.06 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr5_+_175815732 | 0.05 |
ENST00000274787.2
|
HIGD2A
|
HIG1 hypoxia inducible domain family, member 2A |
| chr12_-_56367101 | 0.05 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr1_-_15850676 | 0.05 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr12_+_53835539 | 0.05 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chr7_-_149158187 | 0.05 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr15_+_59664884 | 0.05 |
ENST00000558348.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr12_-_56321649 | 0.05 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr16_-_57513657 | 0.05 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr2_+_158114051 | 0.05 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr18_-_24237339 | 0.05 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr4_+_72052964 | 0.05 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr5_+_173316341 | 0.05 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr12_+_53835383 | 0.05 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr7_+_55433131 | 0.05 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr8_-_133637624 | 0.05 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr12_+_53835425 | 0.05 |
ENST00000549924.1
|
PRR13
|
proline rich 13 |
| chrX_-_30993201 | 0.05 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr15_-_34880646 | 0.05 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr5_-_175815565 | 0.05 |
ENST00000509257.1
ENST00000507413.1 ENST00000510123.1 |
NOP16
|
NOP16 nucleolar protein |
| chr18_-_48351743 | 0.05 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr7_+_73442457 | 0.05 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
| chr5_+_137722255 | 0.05 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr4_-_71532601 | 0.05 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr15_+_59665194 | 0.05 |
ENST00000560394.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr10_-_127505167 | 0.05 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr1_+_100111580 | 0.05 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr1_+_171217622 | 0.05 |
ENST00000433267.1
ENST00000367750.3 |
FMO1
|
flavin containing monooxygenase 1 |
| chr9_-_73483926 | 0.05 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr5_-_34043310 | 0.05 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr11_-_19263145 | 0.05 |
ENST00000532666.1
ENST00000527884.1 |
E2F8
|
E2F transcription factor 8 |
| chr9_-_104500862 | 0.05 |
ENST00000361820.3
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A |
| chr4_-_153274078 | 0.05 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr9_-_16705069 | 0.05 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr1_-_157015162 | 0.05 |
ENST00000368194.3
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr13_-_30160925 | 0.05 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chrX_+_48660107 | 0.05 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr12_-_68845417 | 0.05 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr17_-_21454898 | 0.05 |
ENST00000391411.5
ENST00000412778.3 |
C17orf51
|
chromosome 17 open reading frame 51 |
| chr6_-_34855773 | 0.05 |
ENST00000420584.2
ENST00000361288.4 |
TAF11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr16_-_12184159 | 0.05 |
ENST00000312019.2
|
RP11-276H1.3
|
RP11-276H1.3 |
| chr12_-_109025849 | 0.05 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr6_-_133055896 | 0.05 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr10_+_72238517 | 0.05 |
ENST00000263563.6
|
PALD1
|
phosphatase domain containing, paladin 1 |
| chr11_+_71903169 | 0.05 |
ENST00000393676.3
|
FOLR1
|
folate receptor 1 (adult) |
| chr11_+_65383227 | 0.05 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr11_-_65429891 | 0.05 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr6_-_10412600 | 0.05 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chrX_+_135730297 | 0.04 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr1_-_155271213 | 0.04 |
ENST00000342741.4
|
PKLR
|
pyruvate kinase, liver and RBC |
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.1 | GO:0060067 | cervix development(GO:0060067) |
| 0.0 | 0.1 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:1905174 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.1 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.2 | GO:0021564 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.0 | GO:0018277 | protein deamination(GO:0018277) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.0 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |