Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
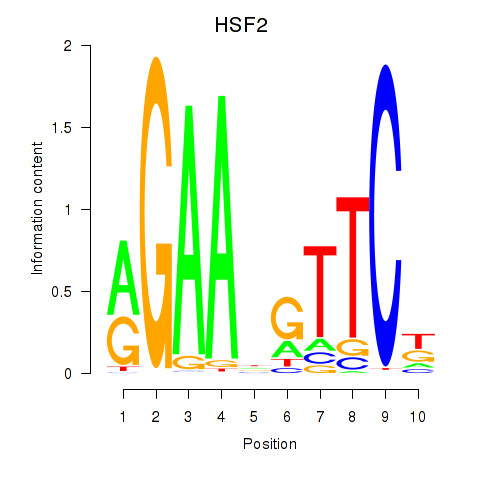
Results for HSF2
Z-value: 0.63
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.8 | heat shock transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF2 | hg19_v2_chr6_+_122720681_122720713 | -0.03 | 9.6e-01 | Click! |
Activity profile of HSF2 motif
Sorted Z-values of HSF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_38261880 | 0.43 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_+_14113592 | 0.42 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr2_+_198365095 | 0.38 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr14_-_75083313 | 0.37 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr1_-_197744763 | 0.36 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr22_+_24577183 | 0.35 |
ENST00000358321.3
|
SUSD2
|
sushi domain containing 2 |
| chr2_+_198365122 | 0.33 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr14_-_102553371 | 0.31 |
ENST00000553585.1
ENST00000216281.8 |
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr11_-_89956461 | 0.29 |
ENST00000320585.6
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr12_-_10007448 | 0.24 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr9_+_33025209 | 0.23 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr2_-_188419200 | 0.23 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr5_+_118691008 | 0.23 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr5_+_56469843 | 0.21 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr11_-_89956227 | 0.21 |
ENST00000457199.2
ENST00000530765.1 |
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr2_-_188419078 | 0.20 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr5_+_56469939 | 0.20 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr6_+_149539767 | 0.20 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr3_-_64673656 | 0.19 |
ENST00000459780.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr4_+_184427235 | 0.19 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chr12_-_123011536 | 0.18 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr2_-_198364581 | 0.18 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr13_-_31736132 | 0.18 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_-_228613026 | 0.17 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr21_-_30445886 | 0.17 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr12_-_14721283 | 0.17 |
ENST00000240617.5
|
PLBD1
|
phospholipase B domain containing 1 |
| chr14_-_52535712 | 0.17 |
ENST00000216286.5
ENST00000541773.1 |
NID2
|
nidogen 2 (osteonidogen) |
| chr16_-_69390209 | 0.17 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr12_-_123011476 | 0.16 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr7_+_120629653 | 0.15 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_+_26240776 | 0.15 |
ENST00000337620.4
|
CBX3
|
chromobox homolog 3 |
| chr5_+_118691706 | 0.15 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chrX_+_135278908 | 0.14 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr12_+_69979210 | 0.14 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr5_+_56469775 | 0.14 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr6_+_32121908 | 0.14 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr16_-_30125177 | 0.13 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr12_+_50794891 | 0.13 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr16_+_55600580 | 0.13 |
ENST00000457326.2
|
CAPNS2
|
calpain, small subunit 2 |
| chr16_+_226658 | 0.13 |
ENST00000320868.5
ENST00000397797.1 |
HBA1
|
hemoglobin, alpha 1 |
| chrX_+_135230712 | 0.13 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr2_+_158733088 | 0.13 |
ENST00000605860.1
|
UPP2
|
uridine phosphorylase 2 |
| chr3_-_129407535 | 0.12 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr12_+_50794947 | 0.12 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr13_-_31736478 | 0.12 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr7_-_33080506 | 0.12 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr7_+_23637763 | 0.12 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr18_-_812231 | 0.11 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr17_+_7608511 | 0.11 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr11_-_82997420 | 0.11 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr16_+_4606347 | 0.11 |
ENST00000444310.4
|
C16orf96
|
chromosome 16 open reading frame 96 |
| chr5_-_95018660 | 0.11 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr7_-_80548493 | 0.10 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr8_-_81083618 | 0.10 |
ENST00000520795.1
|
TPD52
|
tumor protein D52 |
| chr3_-_171489085 | 0.10 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr12_+_69979446 | 0.10 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr6_+_32121789 | 0.09 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr15_-_43785274 | 0.09 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr11_-_82997013 | 0.09 |
ENST00000529073.1
ENST00000529611.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr3_+_177534653 | 0.09 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr10_-_104913367 | 0.09 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr12_+_69979113 | 0.09 |
ENST00000299300.6
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr6_-_53213780 | 0.09 |
ENST00000304434.6
ENST00000370918.4 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr2_-_178128250 | 0.09 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr11_+_86013253 | 0.08 |
ENST00000533986.1
ENST00000278483.3 |
C11orf73
|
chromosome 11 open reading frame 73 |
| chr15_-_93965805 | 0.08 |
ENST00000556708.1
|
RP11-164C12.2
|
RP11-164C12.2 |
| chr18_-_52989525 | 0.08 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr6_+_32121218 | 0.08 |
ENST00000414204.1
ENST00000361568.2 ENST00000395523.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr11_-_111781554 | 0.08 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr11_-_122932730 | 0.08 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr2_+_202937972 | 0.08 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr14_+_101293687 | 0.07 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr13_-_31736027 | 0.07 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr11_-_82997371 | 0.07 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr14_+_93357749 | 0.07 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr1_-_67142710 | 0.07 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr17_-_79817091 | 0.07 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr7_+_30960915 | 0.07 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr2_-_198364552 | 0.07 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr13_+_49684445 | 0.07 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr1_+_151171012 | 0.06 |
ENST00000349792.5
ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chrX_+_106163626 | 0.06 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr11_-_122933043 | 0.06 |
ENST00000534624.1
ENST00000453788.2 ENST00000527387.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr14_+_89060739 | 0.06 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr12_+_58138664 | 0.06 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chrX_-_102942961 | 0.06 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr4_-_159644507 | 0.06 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr6_-_53213587 | 0.06 |
ENST00000542638.1
ENST00000370913.5 ENST00000541407.1 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr19_-_55691377 | 0.06 |
ENST00000589172.1
|
SYT5
|
synaptotagmin V |
| chr7_-_150652924 | 0.06 |
ENST00000330883.4
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr7_+_130126165 | 0.06 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr10_+_135340859 | 0.06 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr6_+_44215603 | 0.06 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr3_-_64673668 | 0.06 |
ENST00000498707.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr5_-_112770674 | 0.06 |
ENST00000390666.3
|
TSSK1B
|
testis-specific serine kinase 1B |
| chr5_-_148442584 | 0.06 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr6_+_151646800 | 0.06 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr16_-_66907139 | 0.05 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr19_+_19516561 | 0.05 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr5_-_148442687 | 0.05 |
ENST00000515425.1
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr18_+_44497455 | 0.05 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr6_-_155635583 | 0.05 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr15_+_96869165 | 0.05 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr9_-_4666421 | 0.05 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr8_-_81083890 | 0.05 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr18_+_3450161 | 0.05 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chrX_+_135279179 | 0.05 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr6_-_135375921 | 0.05 |
ENST00000367820.2
ENST00000314674.3 ENST00000524715.1 ENST00000415177.2 ENST00000367826.2 |
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr6_-_135375986 | 0.05 |
ENST00000525067.1
ENST00000367822.5 ENST00000367837.5 |
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr4_-_74088800 | 0.05 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr7_-_26240357 | 0.05 |
ENST00000354667.4
ENST00000356674.7 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr7_-_103086598 | 0.05 |
ENST00000339444.6
ENST00000356767.4 ENST00000393735.2 ENST00000306312.3 ENST00000432958.2 |
SLC26A5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr13_+_46276441 | 0.05 |
ENST00000310521.1
ENST00000533564.1 |
SPERT
|
spermatid associated |
| chr2_-_10978103 | 0.05 |
ENST00000404824.2
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr6_-_138866823 | 0.05 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr18_+_3449695 | 0.04 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr14_+_103800513 | 0.04 |
ENST00000560338.1
ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr6_-_31782813 | 0.04 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr11_+_1856034 | 0.04 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr7_+_130126012 | 0.04 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr17_-_71410794 | 0.04 |
ENST00000424778.1
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr12_-_45270077 | 0.04 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr4_-_22517620 | 0.04 |
ENST00000502482.1
ENST00000334304.5 |
GPR125
|
G protein-coupled receptor 125 |
| chr18_+_3451584 | 0.04 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_-_112360116 | 0.04 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chrX_-_51239425 | 0.04 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr11_+_117063295 | 0.04 |
ENST00000525478.1
ENST00000532062.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr8_+_134125727 | 0.04 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr19_-_2085323 | 0.04 |
ENST00000591638.1
|
MOB3A
|
MOB kinase activator 3A |
| chr15_-_58357866 | 0.03 |
ENST00000537372.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr17_+_35294075 | 0.03 |
ENST00000254457.5
|
LHX1
|
LIM homeobox 1 |
| chr18_+_3449821 | 0.03 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr20_+_44746885 | 0.03 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chrX_-_102941596 | 0.03 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr5_+_133859996 | 0.03 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chrX_-_102943022 | 0.03 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr20_+_31755934 | 0.03 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr15_+_78556428 | 0.03 |
ENST00000394855.3
ENST00000489435.2 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr7_-_78400364 | 0.03 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_-_123411191 | 0.03 |
ENST00000354792.5
ENST00000508240.1 |
MYLK
|
myosin light chain kinase |
| chr9_-_34589700 | 0.03 |
ENST00000351266.4
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr20_+_4666882 | 0.03 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr2_-_178128199 | 0.03 |
ENST00000449627.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr3_+_177545563 | 0.02 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr8_-_141931287 | 0.02 |
ENST00000517887.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr14_+_32963433 | 0.02 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr5_+_79331164 | 0.02 |
ENST00000350881.2
|
THBS4
|
thrombospondin 4 |
| chr8_-_81083731 | 0.02 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr19_+_54606145 | 0.02 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr19_-_55691472 | 0.02 |
ENST00000537500.1
|
SYT5
|
synaptotagmin V |
| chr3_+_96533621 | 0.02 |
ENST00000542517.1
ENST00000506569.1 |
EPHA6
|
EPH receptor A6 |
| chr11_-_111781610 | 0.02 |
ENST00000525823.1
|
CRYAB
|
crystallin, alpha B |
| chr15_-_70388943 | 0.02 |
ENST00000559048.1
ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr9_-_34589734 | 0.02 |
ENST00000378980.3
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr15_+_78556809 | 0.02 |
ENST00000343789.3
ENST00000394852.3 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr15_-_43785303 | 0.02 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr12_+_54891495 | 0.02 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr17_-_56494882 | 0.02 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr12_-_53012343 | 0.02 |
ENST00000305748.3
|
KRT73
|
keratin 73 |
| chr2_+_219824357 | 0.02 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr8_-_81083341 | 0.02 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chrX_-_72347916 | 0.01 |
ENST00000373518.1
|
NAP1L6
|
nucleosome assembly protein 1-like 6 |
| chr7_-_78400598 | 0.01 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_-_45269430 | 0.01 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr2_+_85581843 | 0.01 |
ENST00000409331.2
ENST00000409013.3 ENST00000409890.2 ENST00000409344.3 ENST00000393852.4 ENST00000428955.2 ENST00000315658.7 ENST00000462891.3 |
ELMOD3
|
ELMO/CED-12 domain containing 3 |
| chr9_+_125795788 | 0.01 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr9_+_841690 | 0.01 |
ENST00000382276.3
|
DMRT1
|
doublesex and mab-3 related transcription factor 1 |
| chr1_-_216596738 | 0.01 |
ENST00000307340.3
ENST00000366943.2 ENST00000366942.3 |
USH2A
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr18_+_59000815 | 0.01 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr14_-_94970559 | 0.01 |
ENST00000556881.1
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr7_-_36764142 | 0.01 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr6_+_152126790 | 0.01 |
ENST00000456483.2
|
ESR1
|
estrogen receptor 1 |
| chr4_-_186877481 | 0.01 |
ENST00000444781.1
ENST00000432655.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_+_1855645 | 0.01 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr12_+_2904102 | 0.01 |
ENST00000001008.4
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr2_-_178128149 | 0.01 |
ENST00000423513.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr6_+_153019069 | 0.01 |
ENST00000532295.1
|
MYCT1
|
myc target 1 |
| chr20_+_58203664 | 0.01 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr4_+_123161120 | 0.01 |
ENST00000446180.1
|
KIAA1109
|
KIAA1109 |
| chr5_-_35089722 | 0.01 |
ENST00000511486.1
ENST00000310101.5 ENST00000231423.3 ENST00000513753.1 ENST00000348262.3 ENST00000397391.3 ENST00000542609.1 |
PRLR
|
prolactin receptor |
| chr1_-_184006611 | 0.01 |
ENST00000546159.1
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr9_+_34989638 | 0.00 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr5_+_148443049 | 0.00 |
ENST00000515304.1
ENST00000507318.1 |
CTC-529P8.1
|
CTC-529P8.1 |
| chr3_-_49131473 | 0.00 |
ENST00000430979.1
ENST00000357496.2 ENST00000437939.1 |
QRICH1
|
glutamine-rich 1 |
| chr18_+_32173276 | 0.00 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr6_+_153019023 | 0.00 |
ENST00000367245.5
ENST00000529453.1 |
MYCT1
|
myc target 1 |
| chr15_-_70388599 | 0.00 |
ENST00000560996.1
ENST00000558201.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chrX_-_101718085 | 0.00 |
ENST00000372750.1
ENST00000372752.1 |
NXF2B
|
nuclear RNA export factor 2B |
| chr15_-_81616446 | 0.00 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr17_-_56494908 | 0.00 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.3 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.2 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.6 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.2 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.3 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.2 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.0 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |