Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HSF1
Z-value: 0.98
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF1
|
ENSG00000185122.6 | heat shock transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg19_v2_chr8_+_145515263_145515299 | 0.22 | 6.7e-01 | Click! |
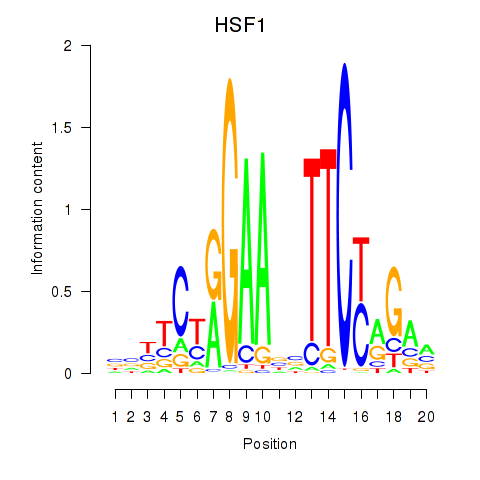
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_139847347 | 0.65 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr19_+_3762703 | 0.59 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr2_-_27851745 | 0.43 |
ENST00000394775.3
ENST00000522876.1 |
CCDC121
|
coiled-coil domain containing 121 |
| chr19_+_8740061 | 0.42 |
ENST00000593792.1
|
CTD-2586B10.1
|
CTD-2586B10.1 |
| chrX_+_49020882 | 0.39 |
ENST00000454342.1
|
MAGIX
|
MAGI family member, X-linked |
| chr19_+_917287 | 0.38 |
ENST00000592648.1
ENST00000234371.5 |
KISS1R
|
KISS1 receptor |
| chr7_-_139763521 | 0.37 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr1_+_35247859 | 0.37 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr19_+_1354930 | 0.35 |
ENST00000591337.1
|
MUM1
|
melanoma associated antigen (mutated) 1 |
| chr17_+_78193443 | 0.34 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr11_-_104840093 | 0.34 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_-_191878162 | 0.34 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr3_+_112930946 | 0.33 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr1_-_1590418 | 0.31 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr11_-_64163297 | 0.31 |
ENST00000457725.1
|
AP003774.6
|
AP003774.6 |
| chr2_-_191878681 | 0.31 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr2_-_191878874 | 0.30 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr16_-_4303767 | 0.30 |
ENST00000573268.1
ENST00000573042.1 |
RP11-95P2.1
|
RP11-95P2.1 |
| chr20_+_36974759 | 0.30 |
ENST00000217407.2
|
LBP
|
lipopolysaccharide binding protein |
| chr19_-_4559814 | 0.29 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr6_+_160693591 | 0.28 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr1_-_206671061 | 0.28 |
ENST00000367119.1
|
C1orf147
|
chromosome 1 open reading frame 147 |
| chr4_+_57396766 | 0.27 |
ENST00000512175.2
|
THEGL
|
theg spermatid protein-like |
| chr11_+_28724129 | 0.27 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr1_+_206138457 | 0.27 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr12_-_108154705 | 0.26 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr19_-_56904799 | 0.26 |
ENST00000589895.1
ENST00000589143.1 ENST00000301310.4 ENST00000586929.1 |
ZNF582
|
zinc finger protein 582 |
| chr14_+_63671105 | 0.26 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr4_-_170192000 | 0.26 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr12_+_50794947 | 0.25 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr12_+_50366620 | 0.24 |
ENST00000315520.5
|
AQP6
|
aquaporin 6, kidney specific |
| chr9_-_34523027 | 0.24 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr1_-_151345159 | 0.24 |
ENST00000458566.1
ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1
|
selenium binding protein 1 |
| chr6_+_33551515 | 0.24 |
ENST00000374458.1
|
GGNBP1
|
gametogenetin binding protein 1 (pseudogene) |
| chr3_-_10334617 | 0.24 |
ENST00000429122.1
ENST00000425479.1 ENST00000335542.8 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr19_+_47813110 | 0.23 |
ENST00000355085.3
|
C5AR1
|
complement component 5a receptor 1 |
| chr11_+_1049862 | 0.23 |
ENST00000534584.1
|
RP13-870H17.3
|
RP13-870H17.3 |
| chr14_+_24407940 | 0.22 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr9_-_34372830 | 0.22 |
ENST00000379142.3
|
KIAA1161
|
KIAA1161 |
| chr7_-_5821225 | 0.21 |
ENST00000416985.1
|
RNF216
|
ring finger protein 216 |
| chr19_-_6720686 | 0.21 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_+_54135310 | 0.21 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chr11_+_72281681 | 0.21 |
ENST00000450804.3
|
RP11-169D4.1
|
RP11-169D4.1 |
| chr19_+_49055332 | 0.21 |
ENST00000201586.2
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr6_+_30294186 | 0.20 |
ENST00000458516.1
|
TRIM39
|
tripartite motif containing 39 |
| chr22_+_48972118 | 0.20 |
ENST00000358295.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr20_-_43753104 | 0.20 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr16_+_30662050 | 0.20 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr17_+_58499844 | 0.20 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr16_-_20338748 | 0.20 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr8_+_58055238 | 0.19 |
ENST00000519314.1
ENST00000519241.1 |
RP11-513O17.2
|
RP11-513O17.2 |
| chr3_-_134092561 | 0.19 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr7_+_13141097 | 0.18 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr2_+_103089756 | 0.18 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr6_-_38607628 | 0.18 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr1_+_92683467 | 0.18 |
ENST00000370375.3
ENST00000370373.2 |
C1orf146
|
chromosome 1 open reading frame 146 |
| chr19_+_5681011 | 0.18 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr19_+_36602104 | 0.18 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr19_-_44258770 | 0.18 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr6_+_168434678 | 0.18 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr19_+_4304585 | 0.18 |
ENST00000221856.6
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr1_-_143913143 | 0.17 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr19_-_53193731 | 0.17 |
ENST00000598536.1
ENST00000594682.2 ENST00000601257.1 |
ZNF83
|
zinc finger protein 83 |
| chr19_-_49567124 | 0.17 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr12_-_10826612 | 0.17 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr2_+_228678550 | 0.17 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr17_+_79859985 | 0.17 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr16_+_28985542 | 0.17 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr19_-_4717835 | 0.16 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr19_+_782755 | 0.16 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr16_-_2770216 | 0.16 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr2_+_109403193 | 0.16 |
ENST00000412964.2
ENST00000295124.4 |
CCDC138
|
coiled-coil domain containing 138 |
| chr3_+_32433363 | 0.16 |
ENST00000465248.1
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr15_+_89584447 | 0.16 |
ENST00000565938.1
|
RP11-326A19.4
|
RP11-326A19.4 |
| chr4_+_166128836 | 0.16 |
ENST00000511305.1
|
KLHL2
|
kelch-like family member 2 |
| chr6_-_170599561 | 0.16 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chrX_+_54947229 | 0.16 |
ENST00000442098.1
ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO
|
trophinin |
| chr1_+_15632231 | 0.16 |
ENST00000375997.4
ENST00000524761.1 ENST00000375995.3 ENST00000401090.2 |
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr19_-_55672037 | 0.16 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr14_-_36536396 | 0.16 |
ENST00000546376.1
|
RP11-116N8.4
|
RP11-116N8.4 |
| chr4_-_122085469 | 0.16 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr2_+_40973618 | 0.15 |
ENST00000420187.1
|
AC007317.1
|
AC007317.1 |
| chr12_-_15374328 | 0.15 |
ENST00000537647.1
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr20_-_23807358 | 0.15 |
ENST00000304725.2
|
CST2
|
cystatin SA |
| chr6_-_41701581 | 0.15 |
ENST00000394283.1
|
TFEB
|
transcription factor EB |
| chr3_-_128690173 | 0.15 |
ENST00000508239.1
|
RP11-723O4.6
|
Uncharacterized protein FLJ43738 |
| chr16_-_12184159 | 0.15 |
ENST00000312019.2
|
RP11-276H1.3
|
RP11-276H1.3 |
| chr19_+_1065922 | 0.15 |
ENST00000539243.2
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr22_+_24577183 | 0.15 |
ENST00000358321.3
|
SUSD2
|
sushi domain containing 2 |
| chr2_+_85981008 | 0.14 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr2_-_11272234 | 0.13 |
ENST00000590207.1
ENST00000417697.2 ENST00000396164.1 ENST00000536743.1 ENST00000544306.1 |
AC062028.1
|
AC062028.1 |
| chr19_+_13842559 | 0.13 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr17_+_18996287 | 0.13 |
ENST00000399091.1
ENST00000443876.1 ENST00000428928.1 |
AC007952.5
|
Uncharacterized protein ENSP00000382042 |
| chr17_+_41005283 | 0.13 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chrX_-_152939252 | 0.13 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr2_-_191885686 | 0.13 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr16_-_3306587 | 0.13 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr14_+_105212297 | 0.13 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr13_-_95248511 | 0.13 |
ENST00000261296.5
|
TGDS
|
TDP-glucose 4,6-dehydratase |
| chr3_+_124223586 | 0.13 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr17_-_79106634 | 0.13 |
ENST00000575363.1
|
AATK
|
apoptosis-associated tyrosine kinase |
| chr14_+_101293687 | 0.13 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr1_+_24118452 | 0.13 |
ENST00000421070.1
|
LYPLA2
|
lysophospholipase II |
| chr4_-_40632757 | 0.13 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr12_-_55042140 | 0.13 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chr9_-_4666421 | 0.12 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr8_+_142402089 | 0.12 |
ENST00000521578.1
ENST00000520105.1 ENST00000523147.1 |
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr5_+_421030 | 0.12 |
ENST00000506456.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr17_-_39041479 | 0.12 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr12_-_9885888 | 0.12 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr19_-_10628117 | 0.12 |
ENST00000333430.4
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr15_+_89921280 | 0.12 |
ENST00000560596.1
ENST00000558692.1 ENST00000538734.2 ENST00000559235.1 |
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr4_-_87515202 | 0.12 |
ENST00000502302.1
ENST00000513186.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_+_161123270 | 0.12 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr17_+_45401308 | 0.12 |
ENST00000331493.2
ENST00000519772.1 ENST00000517484.1 |
EFCAB13
|
EF-hand calcium binding domain 13 |
| chr9_+_6716478 | 0.12 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr7_-_150020814 | 0.12 |
ENST00000477871.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr11_-_107328527 | 0.12 |
ENST00000282251.5
ENST00000433523.1 |
CWF19L2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr2_-_96701722 | 0.12 |
ENST00000434632.1
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr22_-_46644182 | 0.12 |
ENST00000404583.1
ENST00000404744.1 |
CDPF1
|
cysteine-rich, DPF motif domain containing 1 |
| chr3_-_52719888 | 0.12 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr7_-_111032971 | 0.12 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr5_-_141016382 | 0.12 |
ENST00000523088.1
ENST00000305264.3 |
HDAC3
|
histone deacetylase 3 |
| chr4_-_40632844 | 0.12 |
ENST00000505414.1
|
RBM47
|
RNA binding motif protein 47 |
| chr19_-_46974741 | 0.11 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr7_+_116451100 | 0.11 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr17_+_65027509 | 0.11 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr2_+_26624775 | 0.11 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr9_-_116102562 | 0.11 |
ENST00000374193.4
ENST00000465979.1 |
WDR31
|
WD repeat domain 31 |
| chr16_+_57139933 | 0.11 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr11_-_914812 | 0.11 |
ENST00000533059.1
|
CHID1
|
chitinase domain containing 1 |
| chr6_-_161695042 | 0.11 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr19_-_18649181 | 0.11 |
ENST00000596015.1
|
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr6_-_112081113 | 0.11 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr4_-_40632881 | 0.11 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr3_+_32433154 | 0.10 |
ENST00000334983.5
ENST00000349718.4 |
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr6_+_4890226 | 0.10 |
ENST00000343762.5
|
CDYL
|
chromodomain protein, Y-like |
| chr19_+_42806250 | 0.10 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr20_-_23731569 | 0.10 |
ENST00000304749.2
|
CST1
|
cystatin SN |
| chr19_+_49867181 | 0.10 |
ENST00000597546.1
|
DKKL1
|
dickkopf-like 1 |
| chr16_+_57847684 | 0.10 |
ENST00000335616.2
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr19_-_50528392 | 0.10 |
ENST00000600137.1
ENST00000597215.1 |
VRK3
|
vaccinia related kinase 3 |
| chr4_-_8430152 | 0.10 |
ENST00000514423.1
ENST00000503233.1 |
ACOX3
|
acyl-CoA oxidase 3, pristanoyl |
| chr1_+_202830876 | 0.10 |
ENST00000456105.2
|
RP11-480I12.7
|
RP11-480I12.7 |
| chr12_+_34175398 | 0.10 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr7_+_73275483 | 0.10 |
ENST00000320531.2
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28 |
| chr17_+_71228843 | 0.10 |
ENST00000582391.1
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr2_-_139259244 | 0.10 |
ENST00000414911.1
ENST00000431985.1 |
AC097721.2
|
AC097721.2 |
| chr1_-_24469602 | 0.10 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr19_-_12444491 | 0.10 |
ENST00000293725.5
|
ZNF563
|
zinc finger protein 563 |
| chrX_-_74743080 | 0.10 |
ENST00000373367.3
|
ZDHHC15
|
zinc finger, DHHC-type containing 15 |
| chr11_+_33563618 | 0.10 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr17_-_30668887 | 0.10 |
ENST00000581747.1
ENST00000583334.1 ENST00000580558.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr16_-_20339123 | 0.10 |
ENST00000381360.5
|
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr17_+_53828432 | 0.10 |
ENST00000573500.1
|
PCTP
|
phosphatidylcholine transfer protein |
| chr4_+_110834033 | 0.10 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr11_-_102323489 | 0.10 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr10_+_1120312 | 0.10 |
ENST00000436154.1
|
WDR37
|
WD repeat domain 37 |
| chr11_+_94706973 | 0.10 |
ENST00000536741.1
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr19_+_19516561 | 0.10 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr1_-_146054494 | 0.10 |
ENST00000401009.2
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr6_-_128222103 | 0.09 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr1_+_120839005 | 0.09 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr2_+_219247021 | 0.09 |
ENST00000539932.1
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr6_-_41909466 | 0.09 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr12_-_108154925 | 0.09 |
ENST00000228437.5
|
PRDM4
|
PR domain containing 4 |
| chr4_-_84030996 | 0.09 |
ENST00000411416.2
|
PLAC8
|
placenta-specific 8 |
| chr21_-_46221684 | 0.09 |
ENST00000330942.5
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr12_+_4647950 | 0.09 |
ENST00000321524.7
ENST00000543041.1 ENST00000228843.9 ENST00000352618.4 ENST00000544927.1 |
RAD51AP1
|
RAD51 associated protein 1 |
| chr19_-_44952635 | 0.09 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr6_-_41909561 | 0.09 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr13_-_79979919 | 0.09 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr6_+_10694900 | 0.09 |
ENST00000379568.3
|
PAK1IP1
|
PAK1 interacting protein 1 |
| chr22_+_38453207 | 0.09 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr12_+_93772326 | 0.09 |
ENST00000550056.1
ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr5_-_153418407 | 0.09 |
ENST00000522858.1
ENST00000522634.1 ENST00000523705.1 ENST00000524246.1 ENST00000520313.1 ENST00000518102.1 ENST00000351797.4 ENST00000520667.1 ENST00000519808.1 ENST00000522395.1 |
FAM114A2
|
family with sequence similarity 114, member A2 |
| chr22_-_50708781 | 0.09 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr11_+_112046190 | 0.09 |
ENST00000357685.5
ENST00000393032.2 ENST00000361053.4 |
BCO2
|
beta-carotene oxygenase 2 |
| chr9_+_36036430 | 0.09 |
ENST00000377966.3
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr19_-_12945362 | 0.09 |
ENST00000590404.1
ENST00000592204.1 |
RTBDN
|
retbindin |
| chr11_+_94706804 | 0.09 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr7_-_150020750 | 0.09 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr2_-_101034070 | 0.09 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr17_-_4806369 | 0.09 |
ENST00000293780.4
|
CHRNE
|
cholinergic receptor, nicotinic, epsilon (muscle) |
| chr13_+_28527647 | 0.09 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr1_-_204436344 | 0.09 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr12_-_57505121 | 0.09 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr20_-_62168714 | 0.09 |
ENST00000542869.1
|
PTK6
|
protein tyrosine kinase 6 |
| chr2_+_170655789 | 0.09 |
ENST00000409333.1
|
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr8_+_17104401 | 0.09 |
ENST00000324815.3
ENST00000518038.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr1_+_196743912 | 0.09 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr12_-_56321397 | 0.09 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr19_-_2051223 | 0.09 |
ENST00000309340.7
ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr6_+_73331520 | 0.09 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr2_+_102624977 | 0.09 |
ENST00000441002.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chr9_-_19149276 | 0.09 |
ENST00000434144.1
|
PLIN2
|
perilipin 2 |
| chr1_-_149982624 | 0.09 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr7_-_151433342 | 0.08 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr17_+_40714092 | 0.08 |
ENST00000420359.1
ENST00000449624.1 ENST00000585811.1 ENST00000585909.1 ENST00000586771.1 ENST00000421097.2 ENST00000591779.1 ENST00000587858.1 ENST00000587214.1 ENST00000587157.1 ENST00000590958.1 ENST00000393818.2 |
COASY
|
CoA synthase |
| chr5_-_43043272 | 0.08 |
ENST00000314890.3
|
ANXA2R
|
annexin A2 receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.3 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.1 | 0.2 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.2 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.2 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 0.2 | GO:0045360 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.2 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) positive regulation of skeletal muscle tissue growth(GO:0048633) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:1902263 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) L-arginine import(GO:0043091) divalent metal ion export(GO:0070839) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) positive regulation of chondrocyte proliferation(GO:1902732) histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.0 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.0 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.0 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 0.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.2 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 1.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.0 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |