Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
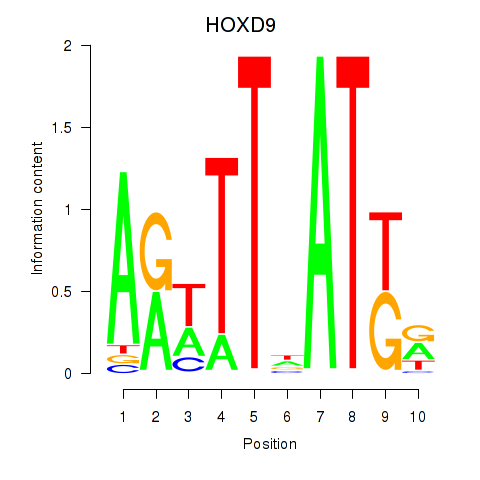
Results for HOXD9
Z-value: 0.47
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.10 | homeobox D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg19_v2_chr2_+_176987088_176987088 | 0.09 | 8.7e-01 | Click! |
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_95616933 | 0.54 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr7_-_35013217 | 0.38 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr1_+_219347203 | 0.30 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr8_-_90993869 | 0.29 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr12_-_91546926 | 0.29 |
ENST00000550758.1
|
DCN
|
decorin |
| chr6_-_18249971 | 0.29 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr1_+_16083098 | 0.27 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr15_+_66797627 | 0.26 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr2_-_55647057 | 0.24 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr3_+_182511266 | 0.24 |
ENST00000323116.5
ENST00000493826.1 |
ATP11B
|
ATPase, class VI, type 11B |
| chr14_+_57671888 | 0.24 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr15_+_66797455 | 0.23 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr15_+_49913175 | 0.22 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr1_+_45140400 | 0.22 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr1_-_241799232 | 0.22 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr15_+_49715449 | 0.21 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr3_-_182703688 | 0.21 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr20_+_5987890 | 0.21 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr2_-_191115229 | 0.21 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr2_+_143635067 | 0.20 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr12_-_90049878 | 0.20 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr10_-_115614127 | 0.20 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr12_+_41136144 | 0.19 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr2_-_170430277 | 0.19 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr13_-_44735393 | 0.19 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr13_+_111748183 | 0.19 |
ENST00000422994.1
|
LINC00368
|
long intergenic non-protein coding RNA 368 |
| chr14_+_45605127 | 0.18 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr13_+_33160553 | 0.17 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr12_-_90049828 | 0.17 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr19_-_19739321 | 0.17 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr3_-_27498235 | 0.17 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr8_-_25281747 | 0.16 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr2_-_55646957 | 0.16 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr14_-_31856397 | 0.16 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr18_-_57027194 | 0.16 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr2_+_145780725 | 0.15 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr2_+_161993465 | 0.15 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr1_+_240177627 | 0.15 |
ENST00000447095.1
|
FMN2
|
formin 2 |
| chr8_+_52730143 | 0.15 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr6_+_18387570 | 0.14 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr12_-_8765446 | 0.14 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr4_+_74606223 | 0.14 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr5_-_96518907 | 0.14 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr2_+_145780739 | 0.14 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chrX_+_123097014 | 0.14 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr14_-_24733444 | 0.14 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chr7_-_92777606 | 0.13 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr14_+_62164340 | 0.13 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr1_+_81771806 | 0.13 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr19_-_53466095 | 0.13 |
ENST00000391786.2
ENST00000434371.2 ENST00000357666.4 ENST00000438970.2 ENST00000270457.4 ENST00000535506.1 ENST00000444460.2 ENST00000457013.2 |
ZNF816
|
zinc finger protein 816 |
| chr4_-_144826682 | 0.13 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr4_-_164534657 | 0.13 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_+_104615595 | 0.13 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr1_+_212475148 | 0.12 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr1_+_45140360 | 0.12 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr2_-_228244013 | 0.12 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr2_-_170430366 | 0.12 |
ENST00000453153.2
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chrY_+_14958970 | 0.12 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr4_-_39640513 | 0.11 |
ENST00000511809.1
ENST00000505729.1 |
SMIM14
|
small integral membrane protein 14 |
| chr7_+_6713376 | 0.11 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr5_+_135496675 | 0.11 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr10_-_94301107 | 0.11 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr14_+_65878650 | 0.11 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr1_-_238108575 | 0.11 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr4_-_36245561 | 0.11 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_+_234627424 | 0.11 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr6_-_10115007 | 0.11 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr7_-_5998714 | 0.11 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr9_-_123812542 | 0.10 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr1_-_100643765 | 0.10 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr17_-_54893250 | 0.10 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr4_-_120225600 | 0.10 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr2_+_145780767 | 0.10 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr6_+_63921351 | 0.10 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr2_+_143635222 | 0.10 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr12_-_15104040 | 0.10 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr11_-_8986474 | 0.10 |
ENST00000525069.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr7_+_120629653 | 0.10 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_-_130353553 | 0.10 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr14_-_38028689 | 0.10 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr7_+_117864708 | 0.10 |
ENST00000357099.4
ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr15_+_57998923 | 0.10 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr5_+_68860949 | 0.10 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr2_+_161993412 | 0.10 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_77469439 | 0.10 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr2_+_210444748 | 0.10 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_+_78383813 | 0.09 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr6_+_131894284 | 0.09 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr1_-_236046872 | 0.09 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr19_+_52800410 | 0.09 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr2_-_231989808 | 0.09 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chrX_-_72434628 | 0.09 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr6_+_24775153 | 0.09 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr17_+_60447579 | 0.09 |
ENST00000450662.2
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr10_+_104613980 | 0.09 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr11_-_104905840 | 0.08 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr1_+_16083123 | 0.08 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr13_+_51483814 | 0.08 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr17_+_61151306 | 0.08 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr4_-_84035868 | 0.08 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr15_-_54025300 | 0.08 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr14_+_20215587 | 0.08 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr2_+_114647504 | 0.08 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr2_+_234668894 | 0.08 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr8_-_117886955 | 0.08 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr8_+_107738343 | 0.08 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr5_+_112043186 | 0.08 |
ENST00000509732.1
ENST00000457016.1 ENST00000507379.1 |
APC
|
adenomatous polyposis coli |
| chr14_-_68000442 | 0.07 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr17_+_58018269 | 0.07 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr12_-_7245152 | 0.07 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr11_-_46639436 | 0.07 |
ENST00000532281.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr2_+_145425573 | 0.07 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr12_+_9144626 | 0.07 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr8_+_86089619 | 0.07 |
ENST00000256117.5
ENST00000416274.2 |
E2F5
|
E2F transcription factor 5, p130-binding |
| chr4_+_25915822 | 0.07 |
ENST00000506197.2
|
SMIM20
|
small integral membrane protein 20 |
| chr10_+_5406935 | 0.07 |
ENST00000380433.3
|
UCN3
|
urocortin 3 |
| chr14_-_81425828 | 0.07 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr11_-_102401469 | 0.07 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chrX_+_12993336 | 0.07 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr4_-_76944621 | 0.06 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chrX_-_77150985 | 0.06 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr5_-_65018834 | 0.06 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr11_+_112047087 | 0.06 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr15_-_45493289 | 0.06 |
ENST00000561278.1
ENST00000290894.8 |
SHF
|
Src homology 2 domain containing F |
| chr14_+_62462541 | 0.06 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr5_-_78808617 | 0.06 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chrY_+_15815447 | 0.06 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chrX_+_118602363 | 0.06 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr5_-_111093167 | 0.06 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr10_-_8095412 | 0.06 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr2_+_54342574 | 0.06 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr7_+_90012986 | 0.06 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr3_-_100712292 | 0.06 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr8_+_101349823 | 0.06 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr2_-_158182105 | 0.05 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr14_+_39583427 | 0.05 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr3_+_107318157 | 0.05 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr3_+_107364683 | 0.05 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr17_+_57233087 | 0.05 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr2_-_225434538 | 0.05 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr14_-_36645674 | 0.05 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr2_-_136288113 | 0.05 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr4_+_186990298 | 0.05 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr3_+_107364769 | 0.05 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr10_-_29084886 | 0.05 |
ENST00000608061.1
ENST00000443246.2 ENST00000446012.1 |
LINC00837
|
long intergenic non-protein coding RNA 837 |
| chrX_-_130423386 | 0.05 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr3_-_116163830 | 0.05 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chrX_-_130423200 | 0.05 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr2_-_136594740 | 0.05 |
ENST00000264162.2
|
LCT
|
lactase |
| chr6_+_149539767 | 0.05 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr1_+_82165350 | 0.05 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr7_+_74379083 | 0.05 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chrX_-_130423240 | 0.05 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr8_-_66701319 | 0.04 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr2_-_118943930 | 0.04 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr6_-_52859968 | 0.04 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr9_-_138391692 | 0.04 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr11_+_69924679 | 0.04 |
ENST00000531604.1
|
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr16_-_18911366 | 0.04 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_-_26231589 | 0.04 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chr17_+_9479944 | 0.04 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr17_+_68071389 | 0.04 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_+_69924639 | 0.04 |
ENST00000538023.1
ENST00000398543.2 |
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr19_+_45843994 | 0.04 |
ENST00000391946.2
|
KLC3
|
kinesin light chain 3 |
| chr14_-_67955426 | 0.03 |
ENST00000554480.1
|
TMEM229B
|
transmembrane protein 229B |
| chr6_-_133055815 | 0.03 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr6_-_39399087 | 0.03 |
ENST00000229913.5
ENST00000541946.1 ENST00000394362.1 |
KIF6
|
kinesin family member 6 |
| chr10_+_115614370 | 0.03 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr1_+_38022513 | 0.03 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr10_+_86184676 | 0.03 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr1_-_48937838 | 0.03 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr4_-_144940477 | 0.03 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr3_-_112564797 | 0.03 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr7_-_107204706 | 0.03 |
ENST00000393603.2
|
COG5
|
component of oligomeric golgi complex 5 |
| chr8_-_80942061 | 0.03 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr18_-_24443151 | 0.03 |
ENST00000440832.3
|
AQP4
|
aquaporin 4 |
| chr4_+_69962212 | 0.03 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr2_-_175712270 | 0.03 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr17_+_68071458 | 0.03 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_-_88974236 | 0.03 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr19_-_56826157 | 0.03 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr13_+_46039037 | 0.03 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr1_-_39339777 | 0.02 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr4_+_71384257 | 0.02 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr3_+_102153859 | 0.02 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr3_+_155860751 | 0.02 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr2_+_210518057 | 0.02 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chrX_+_71401570 | 0.02 |
ENST00000496835.2
ENST00000446576.1 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr12_-_110486348 | 0.02 |
ENST00000547573.1
ENST00000546651.2 ENST00000551185.2 |
C12orf76
|
chromosome 12 open reading frame 76 |
| chr16_+_53483983 | 0.02 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr5_+_61874562 | 0.02 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr11_+_89867803 | 0.02 |
ENST00000321955.4
ENST00000525171.1 ENST00000375944.3 |
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr8_-_10512569 | 0.02 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr15_-_49913126 | 0.02 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr14_+_65878565 | 0.02 |
ENST00000556518.1
ENST00000557164.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr2_-_228497888 | 0.02 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr15_-_90222642 | 0.02 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr6_-_138833630 | 0.02 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr10_-_49459800 | 0.02 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.4 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.4 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |