Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
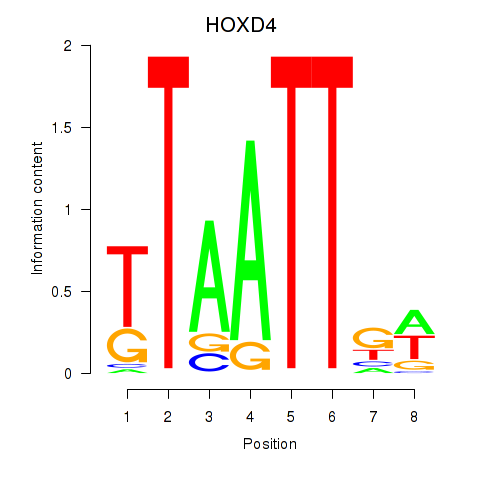
Results for HOXD4
Z-value: 0.56
Transcription factors associated with HOXD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD4
|
ENSG00000170166.5 | homeobox D4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD4 | hg19_v2_chr2_+_177015950_177015950 | -0.51 | 3.0e-01 | Click! |
Activity profile of HOXD4 motif
Sorted Z-values of HOXD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26250835 | 0.51 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr17_+_47448102 | 0.38 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr13_+_51913819 | 0.34 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr8_+_79503458 | 0.32 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr4_+_183065793 | 0.31 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chrX_-_109590174 | 0.29 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr6_-_15548591 | 0.25 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr10_+_69869237 | 0.24 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr18_+_22040593 | 0.24 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr3_-_126327398 | 0.24 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr3_+_142342240 | 0.23 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr3_-_4927447 | 0.22 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr5_-_13944652 | 0.22 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr8_-_40200877 | 0.22 |
ENST00000521030.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr3_+_158787041 | 0.21 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr13_+_110958124 | 0.20 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr4_+_39640787 | 0.20 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr9_-_128246769 | 0.20 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr18_-_21891460 | 0.20 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr5_+_102200948 | 0.20 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_-_73844722 | 0.20 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chrX_+_149867681 | 0.19 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr2_+_179318295 | 0.19 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr18_-_74839891 | 0.19 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr3_-_196987309 | 0.19 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_-_203735484 | 0.19 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr16_-_21663950 | 0.18 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr3_-_191000172 | 0.18 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr22_-_29107919 | 0.17 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr2_+_109237717 | 0.17 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr8_-_18871159 | 0.17 |
ENST00000327040.8
ENST00000440756.2 |
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr10_-_14372870 | 0.16 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chrX_+_139791917 | 0.16 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr4_+_146539415 | 0.16 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr14_-_54425475 | 0.15 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr12_-_118628350 | 0.15 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr12_-_86650077 | 0.15 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_+_117963209 | 0.15 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr1_-_235814048 | 0.14 |
ENST00000450593.1
ENST00000366598.4 |
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4 |
| chr7_+_16793160 | 0.14 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr5_+_59783941 | 0.14 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chrX_+_56590002 | 0.13 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chrX_-_13835147 | 0.13 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr6_-_44281043 | 0.13 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr22_+_23248512 | 0.13 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr18_-_2982869 | 0.13 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr17_+_41132564 | 0.13 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr1_+_67632083 | 0.12 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr1_+_149754227 | 0.12 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr3_+_15045419 | 0.12 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chrX_+_37865804 | 0.11 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr5_-_10761206 | 0.11 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr6_-_53013620 | 0.11 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr4_+_146560245 | 0.11 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr17_-_7218631 | 0.11 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
| chr1_+_202385953 | 0.11 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr3_-_195310802 | 0.11 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr11_-_64684672 | 0.11 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr4_+_169418255 | 0.11 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_-_78450398 | 0.10 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr12_-_39734783 | 0.10 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr7_+_47834908 | 0.10 |
ENST00000418326.2
|
C7orf69
|
chromosome 7 open reading frame 69 |
| chr8_-_42396185 | 0.10 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr14_-_24911868 | 0.10 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr21_+_43823983 | 0.10 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr10_+_95848824 | 0.10 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr12_-_49245936 | 0.10 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr11_+_101983176 | 0.09 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr12_+_112279782 | 0.09 |
ENST00000550735.2
|
MAPKAPK5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr10_+_48189612 | 0.09 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr8_+_24151620 | 0.09 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr4_-_153601136 | 0.09 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr7_+_101460882 | 0.09 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr2_-_165424973 | 0.09 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr14_+_99947715 | 0.08 |
ENST00000389879.5
ENST00000557441.1 ENST00000555049.1 ENST00000555842.1 |
CCNK
|
cyclin K |
| chr12_+_32654965 | 0.08 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_27886330 | 0.08 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr2_+_179317994 | 0.08 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chrX_-_92928557 | 0.08 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr12_+_21168630 | 0.08 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr22_-_30642728 | 0.08 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr2_+_90198535 | 0.08 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr21_-_27423339 | 0.08 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr14_+_23067146 | 0.08 |
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4 |
| chr16_+_103816 | 0.07 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr3_-_4508925 | 0.07 |
ENST00000534863.1
ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1
|
sulfatase modifying factor 1 |
| chr1_-_151762943 | 0.07 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr6_-_136847099 | 0.07 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr4_+_169418195 | 0.07 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_-_159237472 | 0.07 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr16_-_15736953 | 0.07 |
ENST00000548025.1
ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430
|
KIAA0430 |
| chr4_+_169753156 | 0.07 |
ENST00000393726.3
ENST00000507735.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_-_11623804 | 0.07 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr4_+_71019903 | 0.07 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr5_+_101569696 | 0.07 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr12_-_49245916 | 0.07 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr14_-_64761078 | 0.07 |
ENST00000341099.4
ENST00000556275.1 ENST00000542956.1 ENST00000353772.3 ENST00000357782.2 ENST00000267525.6 |
ESR2
|
estrogen receptor 2 (ER beta) |
| chr15_+_64386261 | 0.07 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr1_-_234831899 | 0.07 |
ENST00000442382.1
|
RP4-781K5.6
|
RP4-781K5.6 |
| chr2_-_31361543 | 0.07 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_-_231560790 | 0.06 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr2_-_27579842 | 0.06 |
ENST00000423998.1
ENST00000264720.3 |
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr14_+_29236269 | 0.06 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr4_-_152149033 | 0.06 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr3_+_69812877 | 0.06 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr3_+_159943362 | 0.06 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr4_-_130014705 | 0.06 |
ENST00000503401.1
|
SCLT1
|
sodium channel and clathrin linker 1 |
| chr2_+_110656005 | 0.06 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr15_+_93443419 | 0.06 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr4_+_124571409 | 0.06 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr17_+_66521936 | 0.06 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr8_+_24151553 | 0.06 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr22_+_44427230 | 0.06 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr2_-_203735586 | 0.06 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr3_+_51575596 | 0.06 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_-_186524144 | 0.06 |
ENST00000427785.1
|
RFC4
|
replication factor C (activator 1) 4, 37kDa |
| chr2_+_234826016 | 0.06 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr11_-_72492878 | 0.06 |
ENST00000535054.1
ENST00000545082.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr3_-_137851220 | 0.06 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr18_+_29027696 | 0.06 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr16_+_84801852 | 0.05 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr10_-_61899124 | 0.05 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_-_24911971 | 0.05 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr12_+_41831485 | 0.05 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr5_-_78809950 | 0.05 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chrX_+_36246735 | 0.05 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr1_+_159750776 | 0.05 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr1_-_9953295 | 0.05 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr5_+_111755280 | 0.05 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr17_-_41132410 | 0.05 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr20_+_32150140 | 0.05 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr1_+_203444887 | 0.05 |
ENST00000343110.2
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein |
| chr5_-_55412774 | 0.05 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr12_+_16109519 | 0.04 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr11_+_128563652 | 0.04 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_+_66300464 | 0.04 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr9_+_273038 | 0.04 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr2_-_228497888 | 0.04 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr3_+_140981456 | 0.04 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr14_+_23067166 | 0.04 |
ENST00000216327.6
ENST00000542041.1 |
ABHD4
|
abhydrolase domain containing 4 |
| chr16_-_29910853 | 0.04 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr11_-_36310958 | 0.04 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr9_+_470288 | 0.04 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr2_+_158114051 | 0.04 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr2_-_208030295 | 0.04 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_22138758 | 0.04 |
ENST00000344642.2
ENST00000543870.1 |
LDLRAD2
|
low density lipoprotein receptor class A domain containing 2 |
| chr3_-_157221380 | 0.04 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_-_207024233 | 0.04 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr5_+_95066823 | 0.04 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr2_+_166095898 | 0.04 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr18_-_19283649 | 0.03 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr15_-_64385981 | 0.03 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr17_-_26733179 | 0.03 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr1_+_201617450 | 0.03 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr3_+_197518100 | 0.03 |
ENST00000438796.2
ENST00000414675.2 ENST00000441090.2 ENST00000334859.4 ENST00000425562.2 |
LRCH3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr5_+_66300446 | 0.03 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_+_18629628 | 0.03 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr6_-_36515177 | 0.03 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr15_-_88799661 | 0.03 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr5_+_157158205 | 0.03 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr5_+_69321074 | 0.03 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr2_+_171034646 | 0.03 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr11_-_89224638 | 0.03 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr1_+_214161854 | 0.03 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr11_-_31531121 | 0.02 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr7_+_29186192 | 0.02 |
ENST00000539406.1
|
CHN2
|
chimerin 2 |
| chr6_-_49712072 | 0.02 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr19_-_14064114 | 0.02 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr19_-_5903714 | 0.02 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr12_-_86650045 | 0.02 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_+_80364901 | 0.02 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr2_+_234590556 | 0.02 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr10_+_86184676 | 0.02 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr10_+_13652047 | 0.02 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr9_-_131790550 | 0.02 |
ENST00000372554.4
ENST00000372564.3 |
SH3GLB2
|
SH3-domain GRB2-like endophilin B2 |
| chr6_-_49712091 | 0.02 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr20_+_42136308 | 0.02 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr5_-_39274617 | 0.02 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chr5_-_150727111 | 0.02 |
ENST00000335244.4
ENST00000521967.1 |
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr15_+_36887069 | 0.02 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr1_+_144146808 | 0.02 |
ENST00000369190.5
ENST00000412624.2 ENST00000369365.3 |
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr15_+_55700741 | 0.02 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr14_-_101036119 | 0.02 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr2_+_88047606 | 0.02 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr15_-_65809991 | 0.02 |
ENST00000559526.1
ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8
|
dipeptidyl-peptidase 8 |
| chr5_-_179499086 | 0.02 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr15_+_84115868 | 0.01 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr1_-_151762900 | 0.01 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr6_+_53659746 | 0.01 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr6_+_123317116 | 0.01 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr2_+_33701684 | 0.01 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_+_52083730 | 0.01 |
ENST00000282588.6
ENST00000274311.2 |
ITGA1
PELO
|
integrin, alpha 1 pelota homolog (Drosophila) |
| chr5_+_140743859 | 0.01 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr16_+_24549014 | 0.01 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr13_-_45768841 | 0.01 |
ENST00000379108.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr15_-_64386120 | 0.01 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr4_-_89951028 | 0.01 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr16_+_87636474 | 0.01 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr1_-_92952433 | 0.01 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:2000007 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.0 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.0 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |