Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
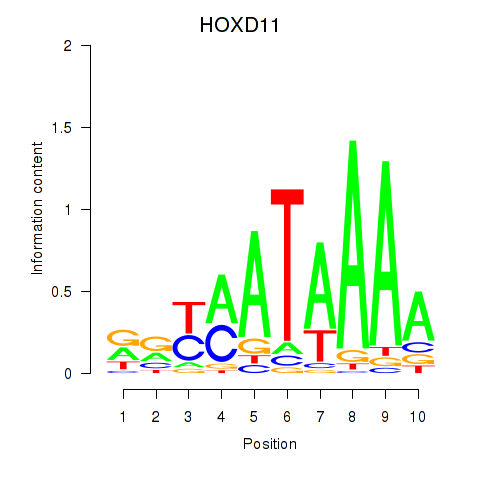
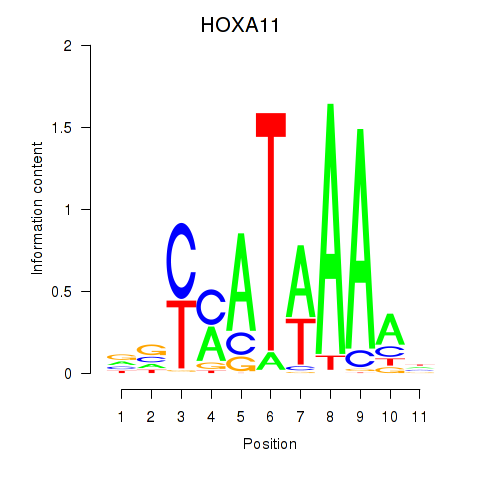
Results for HOXD11_HOXA11
Z-value: 0.78
Transcription factors associated with HOXD11_HOXA11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD11
|
ENSG00000128713.11 | homeobox D11 |
|
HOXA11
|
ENSG00000005073.5 | homeobox A11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA11 | hg19_v2_chr7_-_27224842_27224872 | 0.73 | 1.0e-01 | Click! |
| HOXD11 | hg19_v2_chr2_+_176972000_176972025 | -0.58 | 2.2e-01 | Click! |
Activity profile of HOXD11_HOXA11 motif
Sorted Z-values of HOXD11_HOXA11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_107650219 | 1.14 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr14_+_39944025 | 0.89 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr5_-_159766528 | 0.74 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr12_+_54384370 | 0.57 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr6_-_27775694 | 0.53 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr1_+_202385953 | 0.51 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr2_+_138722028 | 0.49 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr5_+_35852797 | 0.49 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr19_-_10420459 | 0.46 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr10_+_102222798 | 0.44 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr1_-_12679171 | 0.44 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr6_-_121655850 | 0.42 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr13_-_46626820 | 0.42 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr15_+_76352178 | 0.40 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr8_+_101349823 | 0.38 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr12_-_49418407 | 0.37 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr12_+_52056548 | 0.36 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr12_-_112443767 | 0.35 |
ENST00000550233.1
ENST00000546962.1 ENST00000550800.2 |
TMEM116
|
transmembrane protein 116 |
| chr11_-_111649074 | 0.33 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr16_-_21863541 | 0.32 |
ENST00000543654.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr8_-_1922789 | 0.32 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr14_-_25479811 | 0.31 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr3_+_167582561 | 0.31 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr12_-_6579833 | 0.30 |
ENST00000396308.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr6_+_12007963 | 0.30 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr17_+_7211656 | 0.30 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_-_71148413 | 0.29 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr6_-_15548591 | 0.29 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr5_+_121465234 | 0.28 |
ENST00000504912.1
ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr5_-_135290705 | 0.28 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr6_+_26199737 | 0.28 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr16_+_19619083 | 0.28 |
ENST00000538552.1
|
C16orf62
|
chromosome 16 open reading frame 62 |
| chr12_+_6833323 | 0.28 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr18_+_68002675 | 0.28 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr11_+_63606373 | 0.28 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chrX_+_1455668 | 0.28 |
ENST00000432757.1
|
IL3RA
|
interleukin 3 receptor, alpha (low affinity) |
| chr13_-_47471155 | 0.27 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr2_-_160568529 | 0.27 |
ENST00000418770.1
|
AC009961.3
|
AC009961.3 |
| chr11_+_115498761 | 0.27 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chrY_+_15418467 | 0.27 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr12_+_130646999 | 0.26 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr4_+_169633310 | 0.26 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_+_76745385 | 0.26 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr14_-_92247032 | 0.26 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr19_-_45982076 | 0.25 |
ENST00000423698.2
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr16_-_21663950 | 0.25 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr11_-_47447970 | 0.25 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr1_+_62439037 | 0.25 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr2_+_109204743 | 0.25 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr6_+_131958436 | 0.25 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr2_+_97454321 | 0.24 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr9_+_2029019 | 0.24 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_219615984 | 0.24 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr9_+_105757590 | 0.24 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr3_+_140981456 | 0.23 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr10_+_96698406 | 0.23 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr13_-_30951282 | 0.23 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr17_-_46690839 | 0.23 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr7_-_69062391 | 0.23 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr17_+_75369400 | 0.23 |
ENST00000590059.1
|
SEPT9
|
septin 9 |
| chr2_-_228497888 | 0.23 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr2_+_220306238 | 0.23 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr12_-_14924056 | 0.23 |
ENST00000539745.1
|
HIST4H4
|
histone cluster 4, H4 |
| chr12_-_25348007 | 0.23 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr11_-_1606513 | 0.23 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr12_-_62586543 | 0.23 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr1_+_6640108 | 0.23 |
ENST00000377674.4
ENST00000488936.1 |
ZBTB48
|
zinc finger and BTB domain containing 48 |
| chr6_+_42584847 | 0.23 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr3_+_15045419 | 0.23 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr12_+_6833437 | 0.23 |
ENST00000534947.1
ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr1_-_151148442 | 0.23 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr9_-_28670283 | 0.22 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr3_-_112127981 | 0.22 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr10_+_24755416 | 0.22 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr11_+_72983246 | 0.22 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr14_+_105864847 | 0.22 |
ENST00000451127.2
|
TEX22
|
testis expressed 22 |
| chr2_+_158114051 | 0.22 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr5_+_148521454 | 0.22 |
ENST00000508983.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr1_+_229440129 | 0.22 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr11_-_796197 | 0.21 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr10_-_10504285 | 0.21 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr7_+_138915102 | 0.21 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr4_-_85654615 | 0.21 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr16_-_4852915 | 0.21 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr9_-_16705069 | 0.21 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chrX_-_135962923 | 0.21 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr4_+_76439095 | 0.21 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr15_-_85197501 | 0.20 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr2_+_28618532 | 0.20 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr1_-_168464875 | 0.20 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr2_-_111230393 | 0.19 |
ENST00000447537.2
ENST00000413601.2 |
LIMS3L
LIMS3L
|
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein LIM and senescent cell antigen-like domains 3-like |
| chr3_+_169629354 | 0.19 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr8_+_9009202 | 0.19 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr5_-_86534822 | 0.19 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr4_-_84035905 | 0.18 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chrX_+_1455478 | 0.18 |
ENST00000331035.4
|
IL3RA
|
interleukin 3 receptor, alpha (low affinity) |
| chr12_-_71148357 | 0.18 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr20_+_30102231 | 0.18 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr14_+_73706308 | 0.18 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chrX_+_37865804 | 0.18 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr11_-_74204692 | 0.18 |
ENST00000528085.1
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr10_+_127661942 | 0.18 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr7_+_23719749 | 0.18 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr10_+_85933494 | 0.18 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr15_-_101817492 | 0.17 |
ENST00000528346.1
ENST00000531964.1 |
VIMP
|
VCP-interacting membrane protein |
| chr14_-_20774092 | 0.17 |
ENST00000423949.2
ENST00000553828.1 ENST00000258821.3 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr15_-_99789736 | 0.17 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr17_-_39684550 | 0.17 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr6_+_148593425 | 0.17 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr8_-_133772870 | 0.17 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr10_-_90751038 | 0.17 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr16_+_15031300 | 0.17 |
ENST00000328085.6
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr2_+_109204909 | 0.17 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr12_+_70219052 | 0.17 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr9_-_26892765 | 0.17 |
ENST00000520187.1
ENST00000333916.5 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr19_-_10446449 | 0.17 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr10_+_118608998 | 0.17 |
ENST00000409522.1
ENST00000341276.5 ENST00000512864.2 |
ENO4
|
enolase family member 4 |
| chr4_+_124571409 | 0.17 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr12_-_6580094 | 0.17 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr4_-_110723194 | 0.16 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr1_+_92545862 | 0.16 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr4_+_186317133 | 0.16 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr9_+_131085095 | 0.16 |
ENST00000372875.3
|
COQ4
|
coenzyme Q4 |
| chr3_-_9921934 | 0.16 |
ENST00000423850.1
|
CIDEC
|
cell death-inducing DFFA-like effector c |
| chr1_+_174670143 | 0.16 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chrX_-_15332665 | 0.16 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr15_+_67418047 | 0.16 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr12_+_58087738 | 0.16 |
ENST00000552285.1
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr12_+_76653611 | 0.16 |
ENST00000550380.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr19_-_33182616 | 0.16 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr8_+_77318769 | 0.16 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr10_+_47894023 | 0.16 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr3_-_196242233 | 0.16 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_+_150898812 | 0.16 |
ENST00000271640.5
ENST00000448029.1 ENST00000368962.2 ENST00000534805.1 ENST00000368969.4 ENST00000368963.1 ENST00000498193.1 |
SETDB1
|
SET domain, bifurcated 1 |
| chr1_+_181057638 | 0.15 |
ENST00000367577.4
|
IER5
|
immediate early response 5 |
| chr2_-_217236750 | 0.15 |
ENST00000273067.4
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
| chr13_-_24476794 | 0.15 |
ENST00000382140.2
|
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr1_+_154401791 | 0.15 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr6_-_27782548 | 0.15 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr12_+_6833237 | 0.15 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr17_+_61086917 | 0.15 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr13_-_24471194 | 0.15 |
ENST00000382137.3
ENST00000382057.3 |
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr12_+_76653682 | 0.15 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr7_+_79998864 | 0.15 |
ENST00000435819.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_-_61899124 | 0.15 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_+_54230819 | 0.15 |
ENST00000318698.2
ENST00000566473.2 |
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr10_+_135160844 | 0.15 |
ENST00000423766.1
ENST00000458230.1 |
PRAP1
|
proline-rich acidic protein 1 |
| chr10_-_129691195 | 0.14 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr11_-_47447767 | 0.14 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr17_-_27188984 | 0.14 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr5_-_111093759 | 0.14 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr22_-_30642782 | 0.14 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr9_+_116263778 | 0.14 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr7_-_130080977 | 0.14 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr11_+_126173647 | 0.14 |
ENST00000263579.4
|
DCPS
|
decapping enzyme, scavenger |
| chr8_+_94752349 | 0.14 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr5_+_121465207 | 0.14 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr9_-_75488984 | 0.14 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr6_+_158733692 | 0.14 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr2_+_133874577 | 0.14 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr15_-_66797172 | 0.14 |
ENST00000569438.1
ENST00000569696.1 ENST00000307961.6 |
RPL4
|
ribosomal protein L4 |
| chr8_-_38326139 | 0.14 |
ENST00000335922.5
ENST00000532791.1 ENST00000397091.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr4_-_76957214 | 0.14 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr7_+_128784712 | 0.14 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr8_-_38326119 | 0.14 |
ENST00000356207.5
ENST00000326324.6 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr19_-_19626838 | 0.14 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr1_-_89641680 | 0.14 |
ENST00000294671.2
|
GBP7
|
guanylate binding protein 7 |
| chr9_+_101705893 | 0.14 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr12_+_58087901 | 0.14 |
ENST00000315970.7
ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr9_+_131084815 | 0.14 |
ENST00000300452.3
ENST00000609948.1 |
COQ4
|
coenzyme Q4 |
| chrX_+_78003204 | 0.14 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr7_+_73106926 | 0.13 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr5_-_94417186 | 0.13 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr15_+_69857515 | 0.13 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr12_+_43086018 | 0.13 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr17_-_48450534 | 0.13 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr6_-_136788001 | 0.13 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr3_-_52188397 | 0.13 |
ENST00000474012.1
ENST00000296484.2 |
POC1A
|
POC1 centriolar protein A |
| chr9_+_131549610 | 0.13 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr3_+_111260980 | 0.13 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr8_+_77593448 | 0.13 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr13_+_24844857 | 0.13 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr11_+_64001962 | 0.13 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr5_-_36301984 | 0.13 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chrX_+_56590002 | 0.13 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr9_-_98189055 | 0.13 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr19_+_41281060 | 0.13 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr3_-_57326704 | 0.13 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr6_-_131211534 | 0.13 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr9_-_130889990 | 0.13 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr8_-_73793975 | 0.13 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chrX_-_30595959 | 0.12 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr3_-_151176497 | 0.12 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr14_+_56584414 | 0.12 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr16_-_30381580 | 0.12 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr1_+_227127981 | 0.12 |
ENST00000366778.1
ENST00000366777.3 ENST00000458507.2 |
ADCK3
|
aarF domain containing kinase 3 |
| chr11_+_19798964 | 0.12 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr17_+_66521936 | 0.12 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr11_-_65359947 | 0.12 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr1_+_52682052 | 0.12 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD11_HOXA11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.5 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.4 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.2 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.2 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.3 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:1903988 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.0 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.1 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.0 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.0 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.0 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:0033029 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.3 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.3 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0019115 | benzaldehyde dehydrogenase activity(GO:0019115) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.0 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |